

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.6 - phase: 0 /pseudo
(570 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
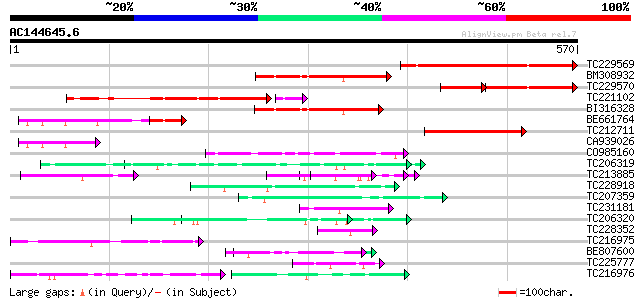
Score E
Sequences producing significant alignments: (bits) Value
TC229569 similar to GB|AAR28016.1|39545906|AY463614 TAF12b {Arab... 286 2e-77
BM308932 191 8e-49
TC229570 weakly similar to UP|TAFC_YEAST (Q03761) Transcription ... 124 4e-48
TC221102 similar to GB|AAR28016.1|39545906|AY463614 TAF12b {Arab... 175 3e-46
BI316328 178 5e-45
BE661764 141 9e-34
TC212711 weakly similar to GB|AAR28015.1|39545904|AY463613 TAF12... 116 2e-26
CA939026 82 8e-16
CO985160 79 4e-15
TC206319 similar to UP|Q9Y0C9 (Q9Y0C9) Ras interacting protein R... 69 7e-12
TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partia... 61 1e-09
TC228918 similar to UP|Q8H6Q7 (Q8H6Q7) CTV.22, partial (4%) 54 2e-07
TC207359 similar to GB|BAB08972.1|9758525|AB024026 auxin respons... 50 2e-06
TC231181 weakly similar to UP|Q86PD9 (Q86PD9) RE65015p, partial ... 49 6e-06
TC206320 similar to UP|Q9W4F9 (Q9W4F9) CG32772-PA, partial (10%) 48 1e-05
TC228352 weakly similar to GB|AAP13389.1|30023712|BT006281 At1g5... 47 2e-05
TC216975 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (28%) 47 2e-05
BE807600 similar to GP|24461867|gb CTV.22 {Poncirus trifoliata},... 46 4e-05
TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 ... 45 9e-05
TC216976 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (6%) 45 9e-05
>TC229569 similar to GB|AAR28016.1|39545906|AY463614 TAF12b {Arabidopsis
thaliana;} , partial (14%)
Length = 853
Score = 286 bits (731), Expect = 2e-77
Identities = 149/179 (83%), Positives = 161/179 (89%), Gaps = 2/179 (1%)
Frame = +3
Query: 394 SGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVT 453
SGATTPGGSSSQGTEA TNQVLGKRKIQDLVAQVDPQG+LDPEVIDLLLE ADDFIDS T
Sbjct: 3 SGATTPGGSSSQGTEA-TNQVLGKRKIQDLVAQVDPQGRLDPEVIDLLLELADDFIDSAT 179
Query: 454 THGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVR 513
THGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKK QS+P N+LHKRRLD VR
Sbjct: 180 THGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKNQSKPQLNDLHKRRLDMVR 359
Query: 514 MLMESSSVPESIVNNSKDISRQG--HPNPAGSHHLMRPLSSDQLVSHSTSSQMLQQMTR 570
LMESSS ES +N+SK++SRQG +P P G+HHL+RPLS +QLVSH+ SQMLQQMTR
Sbjct: 360 TLMESSS-SESSINSSKEMSRQGISNPPPVGTHHLVRPLSPEQLVSHAAGSQMLQQMTR 533
>BM308932
Length = 429
Score = 191 bits (485), Expect = 8e-49
Identities = 111/146 (76%), Positives = 117/146 (80%), Gaps = 9/146 (6%)
Frame = +1
Query: 248 AFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQL 307
AFM+SQLSGLSQNGQP M+ NSLTQ QWLKQMPAMSGPA+PLRLQ HQRQQQ L SS QL
Sbjct: 1 AFMSSQLSGLSQNGQPAMI-NSLTQQQWLKQMPAMSGPAAPLRLQ-HQRQQQ-LGSSTQL 171
Query: 308 QQNSMTLNQQQLSQFMQQQKSMGQPQL------HQQQPSPQQQQQQQL-LQPQQQSQLQA 360
QQNSMTLNQQQLSQ MQQQKSMGQPQL QQQP PQQ QQQ + QPQQQS LQ
Sbjct: 172 QQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQQQQQQQPQPQQLQQQLIQQQPQQQSHLQV 351
Query: 361 SVH--QQQHLHSPRVAGPTGQKSISL 384
SVH QQQ SPR+ GP GQKS+SL
Sbjct: 352 SVHQQQQQQQQSPRMPGPAGQKSLSL 429
Score = 32.3 bits (72), Expect = 0.59
Identities = 35/109 (32%), Positives = 44/109 (40%), Gaps = 3/109 (2%)
Frame = +1
Query: 14 QNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQ 73
Q P+ S P+ P +QQQ S +Q QQ T+N QL Q +Q
Sbjct: 88 QMPAMSGPAAPLRLQHQRQQQLGSS---------TQLQQNSMTLN------QQQLSQLMQ 222
Query: 74 RSPSMSR---LNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
+ SM + L Q Q QQQQ + Q L Q VS QQQ
Sbjct: 223 QQKSMGQPQLLQQQQQQQQQPQPQQLQQQLIQQQPQQQSHLQVSVHQQQ 369
Score = 30.0 bits (66), Expect = 2.9
Identities = 39/140 (27%), Positives = 49/140 (34%)
Frame = +1
Query: 147 PMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRT 206
P +SG AA L +Q+ G Q +S Q L + Q MG P L Q
Sbjct: 94 PAMSGP-AAPLRLQHQRQQQLGSSTQLQQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQ-- 264
Query: 207 NGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMM 266
QQQQ + P QQL QQ + Q Q Q +
Sbjct: 265 ------QQQQQQPQP----QQLQQQLIQQQPQQ----------------------QSHLQ 348
Query: 267 HNSLTQSQWLKQMPAMSGPA 286
+ Q Q +Q P M GPA
Sbjct: 349 VSVHQQQQQQQQSPRMPGPA 408
>TC229570 weakly similar to UP|TAFC_YEAST (Q03761) Transcription initiation
factor TFIID subunit 12 (TBP-associated factor 12)
(TBP-associated factor 61 kDa) (TAFII-61) (TAFII61)
(TAFII-68) (TAFII68), partial (9%)
Length = 676
Score = 124 bits (312), Expect(2) = 4e-48
Identities = 64/94 (68%), Positives = 77/94 (81%), Gaps = 2/94 (2%)
Frame = +3
Query: 479 KNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVRMLMESSSVPESIVNNSKDISRQG-- 536
+NWDL IPGYSSEEKK Q++PL N+LHKRRLD VR LMES ES VN+SK++S QG
Sbjct: 135 ENWDLKIPGYSSEEKKNQNKPLLNDLHKRRLDMVRTLMESLP-SESSVNSSKEMSIQGIS 311
Query: 537 HPNPAGSHHLMRPLSSDQLVSHSTSSQMLQQMTR 570
+P P G+HHL+RPLSS+QL+SH+ SQMLQQMTR
Sbjct: 312 NPPPVGTHHLVRPLSSEQLISHAAGSQMLQQMTR 413
Score = 85.9 bits (211), Expect(2) = 4e-48
Identities = 40/46 (86%), Positives = 43/46 (92%)
Frame = +1
Query: 434 DPEVIDLLLEFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEK 479
+PEVIDLLLE ADDF+DS TTHGC+LAKHRKSSTLES DLLLHLEK
Sbjct: 1 NPEVIDLLLELADDFLDSATTHGCVLAKHRKSSTLESNDLLLHLEK 138
>TC221102 similar to GB|AAR28016.1|39545906|AY463614 TAF12b {Arabidopsis
thaliana;} , partial (15%)
Length = 572
Score = 175 bits (444), Expect(2) = 3e-46
Identities = 112/207 (54%), Positives = 129/207 (62%), Gaps = 1/207 (0%)
Frame = +1
Query: 58 NTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQ 117
+ +NP SN SPS+SRLNQIQ QQ LYGGQM
Sbjct: 1 DNMNPISN-----PCSESPSLSRLNQIQTQQHP--------GLYGGQM------------ 105
Query: 118 QQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSS 177
GQ GHFPML G+G QFNLL+SPRQ GG+VQSSQFSS
Sbjct: 106 -----------------------GQRGHFPMLFGSGT-QFNLLSSPRQ*GGLVQSSQFSS 213
Query: 178 ANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQ-ALNSQ 236
NSAG SLQGM QA+GMMGSPNL SQ+R NG + QQ+R++ Q+RQQLSQQ +LN+
Sbjct: 214 GNSAGPSLQGM-QAMGMMGSPNLTSQLRANGAM-VYAQQLRMSQGQIRQQLSQQGSLNTA 387
Query: 237 QVQGIPRSSSLAFMNSQLSGLSQNGQP 263
QVQ +PRSSSLAFM+SQLSGLSQNGQP
Sbjct: 388 QVQDLPRSSSLAFMSSQLSGLSQNGQP 468
Score = 40.8 bits (94), Expect = 0.002
Identities = 22/34 (64%), Positives = 23/34 (66%)
Frame = +3
Query: 313 TLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQ 346
+L QQQ QQQK MGQPQLHQQQ Q QQQ
Sbjct: 483 SLTQQQ----WQQQKLMGQPQLHQQQQQQQPQQQ 572
Score = 28.5 bits (62), Expect(2) = 3e-46
Identities = 16/32 (50%), Positives = 18/32 (56%)
Frame = +3
Query: 268 NSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQ 299
NSLTQ QW +Q M P + QQ Q QQQ
Sbjct: 480 NSLTQQQW-QQQKLMGQPQLHQQQQQQQPQQQ 572
>BI316328
Length = 407
Score = 178 bits (452), Expect = 5e-45
Identities = 105/138 (76%), Positives = 110/138 (79%), Gaps = 9/138 (6%)
Frame = +2
Query: 247 LAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQ 306
LAFM+SQLSGLSQNGQP M+ NSLTQ QWLKQMPAMSGPA+PLRLQ HQRQQQ L SS Q
Sbjct: 2 LAFMSSQLSGLSQNGQPAMI-NSLTQQQWLKQMPAMSGPAAPLRLQ-HQRQQQ-LGSSTQ 172
Query: 307 LQQNSMTLNQQQLSQFMQQQKSMGQPQL------HQQQPSPQQQQQQQL-LQPQQQSQLQ 359
LQQNSMTLNQQQLSQ MQQQKSMGQPQL QQQP PQQ QQQ + QPQQQS LQ
Sbjct: 173 LQQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQQQQQQQPQPQQLQQQLIQQQPQQQSHLQ 352
Query: 360 ASVH--QQQHLHSPRVAG 375
SVH QQQ SPR+ G
Sbjct: 353 VSVHQQQQQQQQSPRMPG 406
Score = 32.3 bits (72), Expect = 0.59
Identities = 35/109 (32%), Positives = 44/109 (40%), Gaps = 3/109 (2%)
Frame = +2
Query: 14 QNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQ 73
Q P+ S P+ P +QQQ S +Q QQ T+N QL Q +Q
Sbjct: 92 QMPAMSGPAAPLRLQHQRQQQLGSS---------TQLQQNSMTLN------QQQLSQLMQ 226
Query: 74 RSPSMSR---LNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
+ SM + L Q Q QQQQ + Q L Q VS QQQ
Sbjct: 227 QQKSMGQPQLLQQQQQQQQQPQPQQLQQQLIQQQPQQQSHLQVSVHQQQ 373
Score = 29.6 bits (65), Expect = 3.8
Identities = 30/93 (32%), Positives = 37/93 (39%)
Frame = +2
Query: 147 PMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRT 206
P +SG AA L +Q+ G Q +S Q L + Q MG P L Q
Sbjct: 98 PAMSGP-AAPLRLQHQRQQQLGSSTQLQQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQ-- 268
Query: 207 NGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQ 239
QQQQ + P QQL QQ + Q Q
Sbjct: 269 ------QQQQQQPQP----QQLQQQLIQQQPQQ 337
>BE661764
Length = 634
Score = 141 bits (355), Expect = 9e-34
Identities = 93/190 (48%), Positives = 111/190 (57%), Gaps = 22/190 (11%)
Frame = +2
Query: 10 DSQIQNP----SSSNPSIPSPSPISQ----------QQQSPSIHMSNSSPSLSQDQQQL- 54
D QIQNP S+SNP IPSPSP Q Q + S P Q QQ L
Sbjct: 92 DPQIQNPAPSNSNSNPPIPSPSPSHNMASPSLPPLPQDQQQQLQQQLSPPQQQQQQQPLV 271
Query: 55 --HTINTINPNSNFQLQQTLQRSPSMSRLNQIQP-----QQQQQVVARQQAALYGGQMNF 107
+++N +NP SNFQLQQ++QRSPS+SRLNQIQP QQQQ V RQQA LYGGQM+F
Sbjct: 272 SSNSMNNMNPISNFQLQQSMQRSPSLSRLNQIQPQQTTQQQQQFGVMRQQAGLYGGQMSF 451
Query: 108 GGSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKG 167
+ A QQQ GG SNLSRSAL+GQSGHFP G + +L+ SPRQ
Sbjct: 452 AAAGGAGAAQQQQLGG--------SNLSRSALMGQSGHFPCFPG---LERSLIYSPRQXR 598
Query: 168 GMVQSSQFSS 177
+ SS+ +S
Sbjct: 599 KVG*SSRLNS 628
Score = 51.2 bits (121), Expect = 1e-06
Identities = 27/37 (72%), Positives = 29/37 (77%)
Frame = +1
Query: 141 GQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSS 177
G PMLSGAG QFNLL+SP QKGG+VQSSQFSS
Sbjct: 526 GAERALPMLSGAGT-QFNLLSSPXQKGGLVQSSQFSS 633
Score = 37.0 bits (84), Expect = 0.024
Identities = 46/164 (28%), Positives = 57/164 (34%), Gaps = 9/164 (5%)
Frame = +2
Query: 221 PAQMRQQLSQQA-LNSQQVQGIPRSSSLAFMNS-QLSGLSQNGQPGMMHNSLTQSQWLKQ 278
P+ M Q+ A NS IP S M S L L Q+ Q + Q +Q
Sbjct: 80 PSPMDPQIQNPAPSNSNSNPPIPSPSPSHNMASPSLPPLPQDQQQQLQQQLSPPQQQQQQ 259
Query: 279 MPAMSG-------PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQ 331
P +S P S +LQQ ++ L+ Q+Q T QQQ QQ G
Sbjct: 260 QPLVSSNSMNNMNPISNFQLQQSMQRSPSLSRLNQIQPQQTTQQQQQFGVMRQQAGLYGG 439
Query: 332 PQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAG 375
QQQQL S L S Q H P G
Sbjct: 440 QMSFAAAGGAGAAQQQQL----GGSNLSRSALMGQSGHFPCFPG 559
>TC212711 weakly similar to GB|AAR28015.1|39545904|AY463613 TAF12
{Arabidopsis thaliana;} , partial (19%)
Length = 428
Score = 116 bits (291), Expect = 2e-26
Identities = 51/102 (50%), Positives = 78/102 (76%)
Frame = +3
Query: 418 RKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHL 477
R I +LV QVDP KL+PEV D+L++ A++F++S+T GC LAKHRKS+TLE+KD+LLHL
Sbjct: 3 RSIHELVNQVDPLEKLEPEVADILVDIAENFLESITRSGCSLAKHRKSTTLEAKDILLHL 182
Query: 478 EKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVRMLMESS 519
EKNW++T+ G+ ++ K RP ++++HK RL ++ M ++
Sbjct: 183 EKNWNMTLLGFGGDDIKSYRRPTTSDIHKERLTVIKKSMAAT 308
>CA939026
Length = 426
Score = 81.6 bits (200), Expect = 8e-16
Identities = 50/104 (48%), Positives = 59/104 (56%), Gaps = 22/104 (21%)
Frame = +3
Query: 10 DSQIQNP----SSSNPSIPSPSPISQ----------QQQSPSIHMSNSSPSLSQDQQQL- 54
D QIQNP S+SNP IPSPSP Q Q + S P Q QQQ
Sbjct: 93 DPQIQNPAPSNSNSNPPIPSPSPSHNMASPSLPPLPQDQQQQLQQQLSPPQQQQQQQQQQ 272
Query: 55 -------HTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQ 91
+++N +NP SNFQLQQ++QRSPS+SRLNQIQPQQ Q
Sbjct: 273 QQPLVSSNSMNNMNPISNFQLQQSMQRSPSLSRLNQIQPQQTTQ 404
Score = 33.5 bits (75), Expect = 0.26
Identities = 31/88 (35%), Positives = 38/88 (42%), Gaps = 3/88 (3%)
Frame = +3
Query: 282 MSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQL--SQFMQQQKSMGQPQLHQQ-Q 338
M+ P+ P Q Q+Q QQ S Q QQ QQ L S M + QL Q Q
Sbjct: 171 MASPSLPPLPQDQQQQLQQQLSPPQQQQQQQQQQQQPLVSSNSMNNMNPISNFQLQQSMQ 350
Query: 339 PSPQQQQQQQLLQPQQQSQLQASVHQQQ 366
SP + Q+ QPQQ + QQQ
Sbjct: 351 RSPSLSRLNQI-QPQQ------TTQQQQ 413
Score = 32.3 bits (72), Expect = 0.59
Identities = 21/43 (48%), Positives = 22/43 (50%)
Frame = +3
Query: 328 SMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHS 370
+M P L QQQ QQQL PQQQ Q Q QQQ L S
Sbjct: 168 NMASPSLPPLPQDQQQQLQQQLSPPQQQQQQQQ--QQQQPLVS 290
>CO985160
Length = 777
Score = 79.3 bits (194), Expect = 4e-15
Identities = 74/209 (35%), Positives = 95/209 (45%), Gaps = 5/209 (2%)
Frame = -2
Query: 198 PNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGL 257
P L+SQ + + Q QQ++ + Q QQ L Q Q +P+ + Q L
Sbjct: 776 PQLSSQQQQQ--MQQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQ------LQQQQQQL 621
Query: 258 SQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQ 317
Q Q L Q Q +Q+P + +L Q Q+QQQQL QLQQ + QQ
Sbjct: 620 PQLQQQQQQLPQLQQQQ--QQLPQLQQQQQ--QLPQLQQQQQQLP---QLQQQQQQVQQQ 462
Query: 318 QLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPT 377
QL+Q QQQ PQ+ QQQ PQ QQ QL Q QQ QLQ QQQ + V
Sbjct: 461 QLAQLQQQQL----PQIQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQQQQM----VGSGM 306
Query: 378 GQKSISLTG-----SQPDATASGATTPGG 401
GQ + G SQ ++ GAT GG
Sbjct: 305 GQAYVQGPGRSQLVSQGQVSSQGATNIGG 219
Score = 41.2 bits (95), Expect = 0.001
Identities = 36/122 (29%), Positives = 46/122 (37%), Gaps = 4/122 (3%)
Frame = -2
Query: 12 QIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQ----DQQQLHTINTINPNSNFQ 67
Q Q P P QQQQ P + Q QQQL I Q
Sbjct: 572 QQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQVQQQQLAQLQQQQLPQIQQQQQLPQLQ 393
Query: 68 LQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAM 127
LQ+ + +L Q+QPQQQQ V + A G G + + +Q Q S G +
Sbjct: 392 QLPQLQQQQQLPQLQQLQPQQQQMVGSGMGQAYVQGP----GRSQLVSQGQVSSQGATNI 225
Query: 128 GG 129
GG
Sbjct: 224 GG 219
Score = 38.5 bits (88), Expect = 0.008
Identities = 33/100 (33%), Positives = 41/100 (41%), Gaps = 11/100 (11%)
Frame = -2
Query: 12 QIQNPSSSNPSIPSPS--PISQQQQSPSIHMSNSS-PSLSQDQQQLHTINTINPNSNFQL 68
Q+QN P + P QQQQ P + P L Q QQQL + QL
Sbjct: 728 QMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLP-QL 552
Query: 69 QQTLQRSPSMSRLNQIQPQ--------QQQQVVARQQAAL 100
QQ Q+ P + + Q PQ QQQQ+ QQ L
Sbjct: 551 QQQQQQLPQLQQQQQQLPQLQQQQQQVQQQQLAQLQQQQL 432
Score = 35.4 bits (80), Expect = 0.069
Identities = 39/136 (28%), Positives = 53/136 (38%), Gaps = 5/136 (3%)
Frame = -2
Query: 12 QIQNPSSSNPSIPSPS---PISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQL 68
Q+Q P + P QQQQ + L Q QQQ + P Q
Sbjct: 647 QLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQL----PQLQQQQ 480
Query: 69 QQTLQRSPSMSRLNQI-QPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAM 127
QQ Q+ + + Q+ Q QQQQQ+ QQ Q + QQQQ+ G + M
Sbjct: 479 QQVQQQQLAQLQQQQLPQIQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQQQQMVG--SGM 306
Query: 128 GGS-ASNLSRSALIGQ 142
G + RS L+ Q
Sbjct: 305 GQAYVQGPGRSQLVSQ 258
Score = 33.1 bits (74), Expect = 0.34
Identities = 30/92 (32%), Positives = 36/92 (38%), Gaps = 3/92 (3%)
Frame = -2
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQP---Q 87
QQQ + P L Q QQ H P QLQQ Q+ P + + Q P Q
Sbjct: 743 QQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLP----QLQQQQQQLPQLQQQQQQLPQLQQ 576
Query: 88 QQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
QQQQ+ QQ Q+ QQQQ
Sbjct: 575 QQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQ 480
Score = 32.0 bits (71), Expect = 0.77
Identities = 53/220 (24%), Positives = 75/220 (34%), Gaps = 1/220 (0%)
Frame = -2
Query: 48 SQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNF 107
SQ QQQ+ + Q+Q Q P + + Q+ QQQQ+ QQ Q+
Sbjct: 764 SQQQQQMQ-------QQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQL-- 612
Query: 108 GGSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKG 167
QQQQL L Q P L Q L +Q+
Sbjct: 611 ------QQQQQQL----------------PQLQQQQQQLPQLQ---QQQQQLPQLQQQQQ 507
Query: 168 GMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQ-MRQ 226
+ Q Q Q Q QQ + + Q++ L QQQ +L Q +Q
Sbjct: 506 QLPQLQQQQQQVQQQQLAQLQQQQLPQIQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQQQ 327
Query: 227 QLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMM 266
Q+ + VQG RS ++ G + G G M
Sbjct: 326 QMVGSGMGQAYVQGPGRSQLVSQGQVSSQGATNIGGGGFM 207
>TC206319 similar to UP|Q9Y0C9 (Q9Y0C9) Ras interacting protein RIPA, partial
(5%)
Length = 1428
Score = 68.6 bits (166), Expect = 7e-12
Identities = 105/397 (26%), Positives = 152/397 (37%), Gaps = 24/397 (6%)
Frame = +3
Query: 32 QQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQ 91
QQQ+P S + QL +N + NSN L + PS ++ Q QQQQ
Sbjct: 33 QQQNPQFQRS----PMMLGTNQLSHLNPVGQNSNMPLGNHMLNKPSALQMQMFQQQQQQP 200
Query: 92 VVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSG 151
+ R+ M G + + + L G +A MG + G PM S
Sbjct: 201 QMQRKM------MMGLGQAVGMGNLRNNLVG-LAPMGNPMGMGGARGIGGSGISAPMTSI 359
Query: 152 AGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLY 211
AG N+ SP M S + NS Q + ++ S +L S++R ++
Sbjct: 360 AGMG--NMGQSP-----MNLSQTSNITNSISQQFRS--GSLNAAASADLISRLRL---VH 503
Query: 212 TQQQQIRLTPAQMRQQLSQQALNSQQVQ--GIPRSSSLAFMNSQLSGLSQNGQPGMMHNS 269
+ +Q + +P +S ++Q+ P S L N+ + G P MM
Sbjct: 504 SNRQSMLGSPQSNLASIS----GARQIHPGATPSLSMLGRANTMQRPIGPMGPPKMMAG- 668
Query: 270 LTQSQWLKQMPAMSGPASPLRLQQHQR-QQQQLASSGQLQ-QNSMTLNQQQLSQFMQQQK 327
+ ++ Q QQHQ+ QQQQ QLQ Q M QQQL Q QQQ+
Sbjct: 669 --MNLYMSQQ------------QQHQQPQQQQQQHQQQLQLQQHM---QQQLQQQQQQQE 797
Query: 328 -----------------SMGQPQLH---QQQPSPQQQQQQQLLQPQQQSQLQASVHQQQH 367
SMG P L+ QQQ SPQQ Q+ + PQ S ++H
Sbjct: 798 TTSQLQSVVSPPQVGSPSMGVPPLNQQTQQQASPQQMSQRTPMSPQMSS---GAIHAMS- 965
Query: 368 LHSPRVAGPTGQKSISLTGSQPDATASGATTPGGSSS 404
+P + Q S GS T S G + S
Sbjct: 966 AGNPEACPASPQLSSQTLGSVSSITNSPMDMQGVNKS 1076
Score = 43.5 bits (101), Expect = 3e-04
Identities = 68/309 (22%), Positives = 107/309 (34%), Gaps = 6/309 (1%)
Frame = +3
Query: 116 QQQQLSGGVAAMGGSASNLSRSALIGQSGHFP----MLSGAGAAQFNLLTSPRQKGGMVQ 171
QQQ + M + LS +GQ+ + P ML+ A Q + +Q+ M +
Sbjct: 33 QQQNPQFQRSPMMLGTNQLSHLNPVGQNSNMPLGNHMLNKPSALQMQMFQQQQQQPQMQR 212
Query: 172 SSQFSSANSAGQ-SLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQ 230
+ G +L+ + MG+P R GG +T + Q
Sbjct: 213 KMMMGLGQAVGMGNLRNNLVGLAPMGNPMGMGGARGIGGSGISAP---MTSIAGMGNMGQ 383
Query: 231 QALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLR 290
+N Q I S S F + L+ + + + Q + P + +
Sbjct: 384 SPMNLSQTSNITNSISQQFRSGSLNAAASADLISRLRLVHSNRQSMLGSPQSNLASISGA 563
Query: 291 LQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKS-MGQPQLHQQQPSPQQQQQQQL 349
Q H L+ G+ + + M M Q Q HQQ PQQQQQQ
Sbjct: 564 RQIHPGATPSLSMLGRANTMQRPIGPMGPPKMMAGMNLYMSQQQQHQQ---PQQQQQQH- 731
Query: 350 LQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQGTEA 409
Q QLQ H QQ L + T + S+ + S P +Q +
Sbjct: 732 -----QQQLQLQQHMQQQLQQQQQQQETTSQLQSVVSPPQVGSPSMGVPPLNQQTQ--QQ 890
Query: 410 ATNQVLGKR 418
A+ Q + +R
Sbjct: 891 ASPQQMSQR 917
>TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partial (4%)
Length = 540
Score = 61.2 bits (147), Expect = 1e-09
Identities = 49/133 (36%), Positives = 59/133 (43%), Gaps = 12/133 (9%)
Frame = +2
Query: 292 QQHQRQQQQLASSGQLQQNSMT--LNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQL 349
QQHQ+ Q Q QLQQ + QQQL Q QQQ+ PQL QQQ PQ QQQQQL
Sbjct: 17 QQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQL---PQLQQQQQLPQLQQQQQL 187
Query: 350 LQ---PQQQSQLQ-------ASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTP 399
Q PQ Q Q Q + QQQ L + P Q+ + Q G +
Sbjct: 188 QQQQLPQLQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQQQQMVGAGMGQAFGQGPGRSQL 367
Query: 400 GGSSSQGTEAATN 412
++ ATN
Sbjct: 368 VSQGQVSSQGATN 406
Score = 59.7 bits (143), Expect = 3e-09
Identities = 58/152 (38%), Positives = 68/152 (44%), Gaps = 9/152 (5%)
Frame = +2
Query: 259 QNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQ----RQQQQLASSGQLQQNSMTL 314
Q+ Q L Q Q +Q+P M P QQ Q +QQQQL Q QQ L
Sbjct: 20 QHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQQQQLPQLQQQQQ----L 187
Query: 315 NQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQL-----LQPQQQSQLQASVHQQQHLH 369
QQQL Q QQQ QL Q Q PQ QQQQQL LQPQQQ + A + Q
Sbjct: 188 QQQQLPQLQQQQ------QLPQLQQLPQLQQQQQLPQLQQLQPQQQQMVGAGMGQAFG-- 343
Query: 370 SPRVAGPTGQKSISLTGSQPDATASGATTPGG 401
GP + + SQ ++ GAT GG
Sbjct: 344 ----QGPGRSQLV----SQGQVSSQGATNIGG 415
Score = 54.3 bits (129), Expect = 1e-07
Identities = 39/76 (51%), Positives = 42/76 (54%), Gaps = 10/76 (13%)
Frame = +2
Query: 303 SSGQLQQNSMTLNQQQ-LSQFMQQQ-----KSMGQPQLHQQQPSPQQQQQQQLLQPQQQS 356
SS Q QQ+ NQQQ L Q QQQ + PQL QQQ PQ QQQQQL Q QQQ
Sbjct: 2 SSQQQQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQQQQLPQLQQQQ 181
Query: 357 QLQ----ASVHQQQHL 368
QLQ + QQQ L
Sbjct: 182 QLQQQQLPQLQQQQQL 229
Score = 47.8 bits (112), Expect = 1e-05
Identities = 42/133 (31%), Positives = 54/133 (40%), Gaps = 15/133 (11%)
Frame = +2
Query: 12 QIQNPSSSNPSIPSPS--PISQQQQSPSIHMSNSSPSLSQDQQ--QLHTINTINPNSNFQ 67
Q+QN P + P QQQQ P + P L Q QQ QL + Q
Sbjct: 29 QMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQQQQLPQLQQQQQLQQQQLPQ 208
Query: 68 LQQT-----------LQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQ 116
LQQ LQ+ + +L Q+QPQQQQ V A A G G + + +Q
Sbjct: 209 LQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQQQQMVGAGMGQAFGQGP----GRSQLVSQ 376
Query: 117 QQQLSGGVAAMGG 129
Q S G +GG
Sbjct: 377 GQVSSQGATNIGG 415
Score = 36.6 bits (83), Expect = 0.031
Identities = 40/127 (31%), Positives = 49/127 (38%), Gaps = 14/127 (11%)
Frame = +2
Query: 30 SQQQQSPSIHMSNSS---PSLSQDQQQLHTINTINPNSNFQLQ----QTLQRSPSMSRLN 82
S QQQ M N P L Q QQ H P Q Q Q Q+ P + +
Sbjct: 2 SSQQQQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQQQQLPQLQQQQ 181
Query: 83 QIQP------QQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMGGS-ASNLS 135
Q+Q QQQQQ+ QQ Q + QQQQ+ G A MG +
Sbjct: 182 QLQQQQLPQLQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQQQQMVG--AGMGQAFGQGPG 355
Query: 136 RSALIGQ 142
RS L+ Q
Sbjct: 356 RSQLVSQ 376
Score = 32.3 bits (72), Expect = 0.59
Identities = 45/151 (29%), Positives = 55/151 (35%), Gaps = 2/151 (1%)
Frame = +2
Query: 171 QSSQFSSANSAGQSLQGMQQAIGMMGS--PNLASQMRTNGGLYTQQQQIRLTPAQMRQQL 228
Q Q + LQ QQ M P L Q + L QQQ +L Q +QQL
Sbjct: 20 QHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQ----LPQLQQQQQLPQLQQQQQL 187
Query: 229 SQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASP 288
QQ L Q Q QL L Q L Q Q +Q+P
Sbjct: 188 QQQQLPQLQQQ------------QQLPQLQQ----------LPQLQQQQQLP-------- 277
Query: 289 LRLQQHQRQQQQLASSGQLQQNSMTLNQQQL 319
+LQQ Q QQQQ+ +G Q + QL
Sbjct: 278 -QLQQLQPQQQQMVGAGMGQAFGQGPGRSQL 367
>TC228918 similar to UP|Q8H6Q7 (Q8H6Q7) CTV.22, partial (4%)
Length = 1181
Score = 53.5 bits (127), Expect = 2e-07
Identities = 63/227 (27%), Positives = 90/227 (38%), Gaps = 16/227 (7%)
Frame = +2
Query: 182 GQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQ------QQIRLTP-AQMRQQLSQQALN 234
G LQ +G S L Q++ + L +Q Q L P + + Q SQ +
Sbjct: 50 GLVLQSQVHNLGQQHSIPLPGQLQPHQQLLSQNVQNNVASQPNLPPVSSLAQTPSQNIVQ 229
Query: 235 SQQVQGIPRSSSLAFMNSQLSGL------SQNGQPGMMHNSLTQSQWLKQMPAMSGPASP 288
+ +Q I +S+ SQ S L SQ PG + Q +Q
Sbjct: 230 NSNIQNITGPNSVGTTISQNSNLQNMFPGSQRQMPG--RQQIVPPQNSQQYLYQQQFKQK 403
Query: 289 LRLQ-QHQRQQQQLASSGQLQQNSMTLNQQQ--LSQFMQQQKSMGQPQLHQQQPSPQQQQ 345
L+LQ Q Q+QQQ L QLQ + ++ Q + M Q S+ +QQ + QQ
Sbjct: 404 LQLQSQMQQQQQNLLQPNQLQSSQQSIMQTSSVMQPSMMQTSSLSSIPQNQQSTNVQQPM 583
Query: 346 QQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDAT 392
Q PQQ SQ+ Q H P + Q+ L G QP+ T
Sbjct: 584 QS---MPQQHSQVIRHQQQTAVTHQPSL---PSQQQQQLMGHQPNTT 706
Score = 38.5 bits (88), Expect = 0.008
Identities = 59/232 (25%), Positives = 90/232 (38%), Gaps = 5/232 (2%)
Frame = +2
Query: 13 IQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTL 72
+QN +S P++P P+S Q+PS ++ +S ++ TI+ NSN Q
Sbjct: 149 VQNNVASQPNLP---PVSSLAQTPSQNIVQNS-NIQNITGPNSVGTTISQNSNLQ----- 301
Query: 73 QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMGGSAS 132
+M +Q Q +QQ+V Q + Y Q F + +Q QQ
Sbjct: 302 ----NMFPGSQRQMPGRQQIVPPQNSQQYLYQQQFKQKLQLQSQMQQ----------QQQ 439
Query: 133 NLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAI 192
NL + + S Q + TS + M+Q+S SS QS Q
Sbjct: 440 NLLQPNQLQSS-----------QQSIMQTSSVMQPSMMQTSSLSSIPQNQQSTNVQQP-- 580
Query: 193 GMMGSPNLASQMRTNGGLYTQQQQIRLT-----PAQMRQQLSQQALNSQQVQ 239
M P SQ + QQQ +T P+Q +QQL N+ +Q
Sbjct: 581 -MQSMPQQHSQ------VIRHQQQTAVTHQPSLPSQQQQQLMGHQPNTTNMQ 715
>TC207359 similar to GB|BAB08972.1|9758525|AB024026 auxin responsive
transcription factor {Arabidopsis thaliana;} , partial
(25%)
Length = 1263
Score = 50.4 bits (119), Expect = 2e-06
Identities = 60/231 (25%), Positives = 87/231 (36%), Gaps = 21/231 (9%)
Frame = +2
Query: 231 QALNSQQVQGIPRSSSLAFMNSQL------------------SGLSQNGQPGMMHNSLTQ 272
QALNS QG S L +M ++ SGL G +M +
Sbjct: 347 QALNSLNFQG---SGLLPWMQQRMDPTLLANDHNQQYQAMFASGLQNLGSGDLMRQQIMN 517
Query: 273 SQWLKQMPAMSG-PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQ 331
Q SG P PL+LQQ Q QQ ++S+ LQ + + + +Q+ + +
Sbjct: 518 FQQPFNYLQQSGNPNPPLQLQQPQAIQQSVSSNNILQPQAQVMAENLSQHLLQKSHNNRE 697
Query: 332 PQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQ--P 389
Q QQQ Q LLQ Q +HQ+QH P P+ K L S P
Sbjct: 698 DQTQQQQQQRHTYQDTVLLQSDQ-------LHQRQHSGLP---SPSYSKPDFLDSSMKFP 847
Query: 390 DATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDL 440
+ + G G +G+ N R Q ++ + PQ P+ L
Sbjct: 848 ASVSPGQNMLGSLCPEGSGNLLNL---SRSSQSMLTEQLPQQSWAPKFTPL 991
Score = 30.4 bits (67), Expect = 2.2
Identities = 42/156 (26%), Positives = 64/156 (40%), Gaps = 2/156 (1%)
Frame = +2
Query: 61 NPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL 120
NPN QLQQ S+S N +QPQ Q Q L Q + + QQQQ
Sbjct: 554 NPNPPLQLQQPQAIQQSVSSNNILQPQAQVMAENLSQHLL---QKSHNNREDQTQQQQQ- 721
Query: 121 SGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPR-QKGGMVQSS-QFSSA 178
+ + ++ QS +G L SP K + SS +F ++
Sbjct: 722 ----------QRHTYQDTVLLQSDQLHQRQHSG------LPSPSYSKPDFLDSSMKFPAS 853
Query: 179 NSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQ 214
S GQ++ G ++ GS NL + R++ + T+Q
Sbjct: 854 VSPGQNMLG---SLCPEGSGNLLNLSRSSQSMLTEQ 952
Score = 29.6 bits (65), Expect = 3.8
Identities = 46/208 (22%), Positives = 77/208 (36%), Gaps = 18/208 (8%)
Frame = +2
Query: 117 QQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFS 176
Q + G+ +G + +L R ++ F L +G +P + Q
Sbjct: 449 QAMFASGLQNLG--SGDLMRQQIMNFQQPFNYLQQSG--------NPNPPLQLQQPQAIQ 598
Query: 177 SANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQ 236
+ S+ LQ Q + S +L + N TQQQQ + Q L L+ +
Sbjct: 599 QSVSSNNILQPQAQVMAENLSQHLLQKSHNNREDQTQQQQQQRHTYQDTVLLQSDQLHQR 778
Query: 237 QVQGIPR---------SSSLAFMNS------QLSGLSQNGQPGMMHNS-LTQSQWLKQMP 280
Q G+P SS+ F S L L G +++ S +QS +Q+P
Sbjct: 779 QHSGLPSPSYSKPDFLDSSMKFPASVSPGQNMLGSLCPEGSGNLLNLSRSSQSMLTEQLP 958
Query: 281 AMSGPA--SPLRLQQHQRQQQQLASSGQ 306
S +PL++ Q + SG+
Sbjct: 959 QQSWAPKFTPLQINAFGNSMQHVQYSGK 1042
>TC231181 weakly similar to UP|Q86PD9 (Q86PD9) RE65015p, partial (10%)
Length = 671
Score = 48.9 bits (115), Expect = 6e-06
Identities = 41/104 (39%), Positives = 51/104 (48%), Gaps = 9/104 (8%)
Frame = +2
Query: 292 QQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMG------QPQLHQQQPSPQQQQ 345
Q +Q+QQQQL QLQQ+ + L QQ Q + QQ+ QPQ QQQ QQQQ
Sbjct: 245 QYYQQQQQQL----QLQQHYIQLQQQSFQQGLSQQQLQHSLLEPLQPQQLQQQVLQQQQQ 412
Query: 346 QQ-QLLQPQQQSQLQASVHQQQ--HLHSPRVAGPTGQKSISLTG 386
Q Q QP QQ + + QQQ S GQ ++L G
Sbjct: 413 QYFQQQQPLQQEHPENLMQQQQPSTQSSSHPVADQGQAIVTLQG 544
Score = 33.9 bits (76), Expect = 0.20
Identities = 37/126 (29%), Positives = 48/126 (37%), Gaps = 11/126 (8%)
Frame = +2
Query: 249 FMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQ----QQLASS 304
+ N Q +S M N+ + +Q + ++LQQ QQ QQL S
Sbjct: 173 YYNMQCKAMSDMDICRMTKNNQLWQYYQQQQQQLQLQQHYIQLQQQSFQQGLSQQQLQHS 352
Query: 305 -------GQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQ 357
QLQQ L QQQ F QQQ P QQ+ + L QQQ
Sbjct: 353 LLEPLQPQQLQQQ--VLQQQQQQYFQQQQ--------------PLQQEHPENLMQQQQPS 484
Query: 358 LQASVH 363
Q+S H
Sbjct: 485 TQSSSH 502
Score = 31.2 bits (69), Expect = 1.3
Identities = 35/125 (28%), Positives = 46/125 (36%), Gaps = 2/125 (1%)
Frame = +2
Query: 213 QQQQIRLTP--AQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSL 270
QQQQ++L Q++QQ QQ L+ QQ+Q +SL
Sbjct: 260 QQQQLQLQQHYIQLQQQSFQQGLSQQQLQ----------------------------HSL 355
Query: 271 TQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMG 330
+ P P +LQQ QQQQ Q Q L Q+ MQQQ+
Sbjct: 356 LE------------PLQPQQLQQQVLQQQQ----QQYFQQQQPLQQEHPENLMQQQQPST 487
Query: 331 QPQLH 335
Q H
Sbjct: 488 QSSSH 502
>TC206320 similar to UP|Q9W4F9 (Q9W4F9) CG32772-PA, partial (10%)
Length = 1120
Score = 48.1 bits (113), Expect = 1e-05
Identities = 66/256 (25%), Positives = 95/256 (36%), Gaps = 24/256 (9%)
Frame = +2
Query: 173 SQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQA 232
+ + + GQ+ + Q + S SQ +G + L+ ++ Q Q
Sbjct: 5 TSIAGMGNMGQNPMNLSQTSNITNS---ISQQFRSGSINAAASADLLSKLRLVHQNRQGM 175
Query: 233 LNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQ 292
L S Q S + LS G+ M + K M M+ S + Q
Sbjct: 176 LGSSQSNIASISGARQIHPGGTPSLSMLGRANTMQRPIGPMGPPKIMAGMNLYMSQQQQQ 355
Query: 293 QHQR-----QQQQLASSGQLQQNSMTLNQQQLSQFMQQQK----------------SMGQ 331
QHQ+ QQQQ QLQQ+ QQQL Q QQ+ SMG
Sbjct: 356 QHQQPQPQQQQQQHQQQLQLQQHM----QQQLQQQQQQETTSQLQAVVSPPQVGSPSMGI 523
Query: 332 PQLHQQ---QPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQ 388
P ++QQ Q SPQQ Q+ + PQ S ++H + +P + Q S GS
Sbjct: 524 PPMNQQAQQQASPQQMSQRTPMSPQMSS---GAIH-AMNAGNPEACPASPQLSSQTLGSV 691
Query: 389 PDATASGATTPGGSSS 404
T S G + S
Sbjct: 692 SSITNSPMDMQGVNKS 739
Score = 47.8 bits (112), Expect = 1e-05
Identities = 64/243 (26%), Positives = 94/243 (38%), Gaps = 21/243 (8%)
Frame = +2
Query: 123 GVAAMGGSASNLSRSALIGQSGHFPMLSGA--GAAQFNLLTSPR----QKGGMVQSSQFS 176
G+ MG + NLS+++ I S SG+ AA +LL+ R + GM+ SSQ +
Sbjct: 17 GMGNMGQNPMNLSQTSNITNSISQQFRSGSINAAASADLLSKLRLVHQNRQGMLGSSQSN 196
Query: 177 SANSAGQ-----------SLQG----MQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTP 221
A+ +G S+ G MQ+ IG MG P + + M QQQ + P
Sbjct: 197 IASISGARQIHPGGTPSLSMLGRANTMQRPIGPMGPPKIMAGMNLYMSQQQQQQHQQPQP 376
Query: 222 AQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPA 281
Q +QQ QQ Q +Q Q Q + +Q Q + P
Sbjct: 377 QQQQQQHQQQLQLQQHMQ-------------------QQLQQQQQQETTSQLQAVVSPPQ 499
Query: 282 MSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSP 341
+ P+ + Q QQQ AS Q+ Q T Q+S + G P+ P
Sbjct: 500 VGSPSMGIPPMNQQAQQQ--ASPQQMSQR--TPMSPQMSSGAIHAMNAGNPEACPASPQL 667
Query: 342 QQQ 344
Q
Sbjct: 668 SSQ 676
Score = 39.3 bits (90), Expect = 0.005
Identities = 28/88 (31%), Positives = 38/88 (42%)
Frame = +2
Query: 331 QPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPD 390
Q Q QQP PQQQQQQ Q QLQ H QQ L + T Q ++ Q
Sbjct: 344 QQQQQHQQPQPQQQQQQH------QQQLQLQQHMQQQLQQQQQQETTSQLQAVVSPPQVG 505
Query: 391 ATASGATTPGGSSSQGTEAATNQVLGKR 418
+ + G + Q + A+ Q + +R
Sbjct: 506 SPSMGIPP---MNQQAQQQASPQQMSQR 580
>TC228352 weakly similar to GB|AAP13389.1|30023712|BT006281 At1g55080
{Arabidopsis thaliana;} , partial (50%)
Length = 1007
Score = 47.4 bits (111), Expect = 2e-05
Identities = 28/64 (43%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Frame = +3
Query: 310 NSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSP----QQQQQQQLLQPQQQSQLQASVHQQ 365
NS + + Q + F+QQQ+ Q Q QQ +P QQQQQQ+LLQ QQQ Q ++HQ
Sbjct: 195 NSQSQSNQDPNLFLQQQQQFLQSQPFQQALAPQSPFQQQQQQRLLQQQQQQQQPQNLHQS 374
Query: 366 QHLH 369
H
Sbjct: 375 LASH 386
Score = 30.0 bits (66), Expect = 2.9
Identities = 20/49 (40%), Positives = 24/49 (48%), Gaps = 4/49 (8%)
Frame = +3
Query: 296 RQQQQLASSGQLQQN---SMTLNQQQLSQFMQQQKSMGQPQ-LHQQQPS 340
+QQQQ S QQ QQQ + +QQQ+ QPQ LHQ S
Sbjct: 237 QQQQQFLQSQPFQQALAPQSPFQQQQQQRLLQQQQQQQQPQNLHQSLAS 383
>TC216975 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (28%)
Length = 1544
Score = 47.4 bits (111), Expect = 2e-05
Identities = 51/201 (25%), Positives = 88/201 (43%), Gaps = 6/201 (2%)
Frame = +1
Query: 1 MAENVIGSPDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTI 60
M +N + S +Q SS + QSPS + + P+L Q +++
Sbjct: 631 MRQNSMNSSPGSLQREGSS---------FNNSNQSPSSALQGAGPALIPGPMQNSSVSGF 783
Query: 61 -NPNSNFQLQQTLQRSPSMSR---LNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQ 116
+P Q QQ + PS+S L Q Q Q A QQ ++ + +
Sbjct: 784 PSPRLPPQQQQHHLQQPSLSANALLQQNHSQGSQGNQALQQQMIH----------QLLQE 933
Query: 117 QQQLSGGVA--AMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQ 174
+GGV ++GG ++N++++AL G GH+P LSG G+A P + +++
Sbjct: 934 MSNNNGGVQPQSLGGPSANMAKNAL-GFGGHYPSLSG-GSANVTGNNGPMSRNNSFKTTA 1107
Query: 175 FSSANSAGQSLQGMQQAIGMM 195
S +++AG + G+ Q M
Sbjct: 1108NSDSSAAGGN-NGLNQRTSEM 1167
Score = 40.8 bits (94), Expect = 0.002
Identities = 35/120 (29%), Positives = 50/120 (41%), Gaps = 10/120 (8%)
Frame = +1
Query: 224 MRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMS 283
MRQ + S Q +G ++S +S L G PG M NS +
Sbjct: 631 MRQNSMNSSPGSLQREGSSFNNSNQSPSSALQGAGPALIPGPMQNS--------SVSGFP 786
Query: 284 GPASPLRLQQHQRQQQQLASSGQLQQN-------SMTLNQQQLSQFMQQQKSMG---QPQ 333
P P + QQH QQ L+++ LQQN + L QQ + Q +Q+ + QPQ
Sbjct: 787 SPRLPPQQQQHHLQQPSLSANALLQQNHSQGSQGNQALQQQMIHQLLQEMSNNNGGVQPQ 966
Score = 30.4 bits (67), Expect = 2.2
Identities = 46/173 (26%), Positives = 69/173 (39%), Gaps = 13/173 (7%)
Frame = +1
Query: 208 GGLYTQQQQIRLTPAQMRQQLSQQALNSQ-QVQGIPRSSSLAFMNSQLSGLSQNGQPGMM 266
G + + + RL A Q Q + VQ +P + ++L L+ PG+
Sbjct: 376 GAIESLKNYPRLATASKHQMQKMQEMEQLGNVQCLPTDQNTL---NKLMALN----PGL- 531
Query: 267 HNSLTQSQWLKQMPAMSGPA-SPLRLQQHQ----RQQQQLASSGQLQQNSMTLNQ--QQL 319
+N + S + A+SG A + L L +Q RQ +S G LQ+ + N Q
Sbjct: 532 NNHINNSHNMVNRGALSGSAQAALALNNYQNLLMRQNSMNSSPGSLQREGSSFNNSNQSP 711
Query: 320 SQFMQQQKSMGQPQLHQQQ-----PSPQQQQQQQLLQPQQQSQLQASVHQQQH 367
S +Q P Q PSP+ QQQ QQ S ++ QQ H
Sbjct: 712 SSALQGAGPALIPGPMQNSSVSGFPSPRLPPQQQQHHLQQPSLSANALLQQNH 870
>BE807600 similar to GP|24461867|gb CTV.22 {Poncirus trifoliata}, partial
(4%)
Length = 414
Score = 46.2 bits (108), Expect = 4e-05
Identities = 40/143 (27%), Positives = 57/143 (38%)
Frame = +3
Query: 226 QQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGP 285
Q ++Q L Q +Q S + + + QN P M + +P +
Sbjct: 21 QPPNRQQLLPQNIQNSIASQPSNIAQAPIQNVGQNN-PNMQN-----------IPGQNSV 164
Query: 286 ASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQ 345
S + Q Q S Q+Q + QQQ Q Q+ + Q Q+ Q + QQ
Sbjct: 165 GSTIS-QNSNMQNMFPGSQRQIQGRQQVVPQQQQQQSQNSQQYIYQQQMQHQLLRQKLQQ 341
Query: 346 QQQLLQPQQQSQLQASVHQQQHL 368
QQQ Q QQQ Q Q QQQ+L
Sbjct: 342 QQQQQQQQQQQQQQQQQQQQQNL 410
Score = 45.8 bits (107), Expect = 5e-05
Identities = 46/147 (31%), Positives = 62/147 (41%), Gaps = 6/147 (4%)
Frame = +3
Query: 218 RLTPAQMRQQLSQQALNSQQV------QGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLT 271
+L P ++ ++ Q N Q Q P ++ NS S +SQN M N
Sbjct: 39 QLLPQNIQNSIASQPSNIAQAPIQNVGQNNPNMQNIPGQNSVGSTISQNSN---MQNMFP 209
Query: 272 QSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQ 331
SQ +Q+ R Q +QQQQ + + Q + Q L Q +QQQ
Sbjct: 210 GSQ--RQIQG--------RQQVVPQQQQQQSQNSQQYIYQQQMQHQLLRQKLQQQ----- 344
Query: 332 PQLHQQQPSPQQQQQQQLLQPQQQSQL 358
QQQ QQQQQQQ Q QQQ+ L
Sbjct: 345 ----QQQQQQQQQQQQQQQQQQQQNLL 413
Score = 37.0 bits (84), Expect = 0.024
Identities = 32/107 (29%), Positives = 46/107 (42%)
Frame = +3
Query: 13 IQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTL 72
IQN +S PS + +PI ++ ++P++ Q +TI+ NSN Q
Sbjct: 57 IQNSIASQPSNIAQAPIQ--------NVGQNNPNMQNIPGQNSVGSTISQNSNMQNMFPG 212
Query: 73 QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
+ R Q+ PQQQQQ Q +Y QM QQQQ
Sbjct: 213 SQRQIQGR-QQVVPQQQQQQSQNSQQYIYQQQMQHQLLRQKLQQQQQ 350
Score = 28.5 bits (62), Expect = 8.5
Identities = 28/98 (28%), Positives = 39/98 (39%), Gaps = 13/98 (13%)
Frame = +3
Query: 13 IQNPSSSNPS---IPSPSPISQQQQSPSIHMSNSSPSLSQD----------QQQLHTINT 59
IQN +NP+ IP + + S +M N P + QQQ + N+
Sbjct: 105 IQNVGQNNPNMQNIPGQNSVGSTISQNS-NMQNMFPGSQRQIQGRQQVVPQQQQQQSQNS 281
Query: 60 INPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQ 97
Q+Q L R + Q Q QQQQQ +QQ
Sbjct: 282 QQYIYQQQMQHQLLRQKLQQQQQQQQQQQQQQQQQQQQ 395
>TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 protein
(Corresponding sequence ZK643.8), partial (7%)
Length = 762
Score = 45.1 bits (105), Expect = 9e-05
Identities = 42/102 (41%), Positives = 43/102 (41%), Gaps = 10/102 (9%)
Frame = +3
Query: 285 PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQ----FMQQQKSMGQPQLHQQQPS 340
P P QQ Q QQQL Q Q L QQL F Q PQ H QPS
Sbjct: 150 PPQPPVQQQQQPPQQQLQQPPQ-QPPQQHLAPQQLPPQPPIFQQTYPPFHLPQFHHHQPS 326
Query: 341 PQQQQQQQLLQPQQ------QSQLQASVHQQQHLHSPRVAGP 376
Q QQQQQ QP Q Q Q Q HQQ H P +GP
Sbjct: 327 LQFQQQQQ--QPPQPPPQLFQHQAQPPSHQQFH---P*ESGP 437
Score = 32.3 bits (72), Expect = 0.59
Identities = 22/71 (30%), Positives = 33/71 (45%), Gaps = 6/71 (8%)
Frame = +3
Query: 291 LQQHQRQQQQLASSGQLQQ----NSMTLNQQQLSQFMQQQKSMGQPQ--LHQQQPSPQQQ 344
L+ H + + + +Q+ +M LN + + Q + PQ + QQQ PQQQ
Sbjct: 18 LKPHFNHESPVTAIAAIQELEVLGTMDLNVPLRDETLPHQVELPPPQPPVQQQQQPPQQQ 197
Query: 345 QQQQLLQPQQQ 355
QQ QP QQ
Sbjct: 198 LQQPPQQPPQQ 230
>TC216976 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (6%)
Length = 1015
Score = 45.1 bits (105), Expect = 9e-05
Identities = 66/227 (29%), Positives = 93/227 (40%), Gaps = 10/227 (4%)
Frame = +2
Query: 1 MAENVIGSPDSQIQNPSSS-NPSIPSPSPISQQQQSPSI---HMSNS------SPSLSQD 50
M +N + S +Q SS N S PSPS + Q P++ M NS SP L+
Sbjct: 104 MRQNSMNSSPGSLQREGSSFNNSNPSPSS-ALQGTGPALIPGSMQNSPVGGFPSPHLTPQ 280
Query: 51 QQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGS 110
QQQ + ++N LQQ + + Q QQQQ++ Q L N GG
Sbjct: 281 QQQQQLLQQRTLSANGLLQQNHSQGSQGN-----QALQQQQMI---QQLLQEMSNNNGG- 433
Query: 111 AAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMV 170
Q Q GG A G N+S++ + G GH P LSG A P +
Sbjct: 434 -----LQSQSLGGHNANG----NISKNTM-GFGGHTPSLSGGSA------NVPGNNRPIS 565
Query: 171 QSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQI 217
+++ F +A+++ S G G G S M+ N L Q I
Sbjct: 566 RNNSFKTASNSDSSAAG-----GNNGFNQRTSDMQQNLHLQDVAQDI 691
Score = 43.5 bits (101), Expect = 3e-04
Identities = 50/182 (27%), Positives = 67/182 (36%), Gaps = 3/182 (1%)
Frame = +2
Query: 224 MRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMS 283
MRQ + S Q +G ++S +S L G PG M NS P
Sbjct: 104 MRQNSMNSSPGSLQREGSSFNNSNPSPSSALQGTGPALIPGSMQNS----------PVGG 253
Query: 284 GPASPLRLQQHQRQ---QQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPS 340
P+ L QQ Q+Q Q+ L+++G LQQN Q S G L
Sbjct: 254 FPSPHLTPQQQQQQLLQQRTLSANGLLQQN-------------HSQGSQGNQALQ----- 379
Query: 341 PQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPG 400
QQQ QQLLQ + Q Q L G + ++ G P + A PG
Sbjct: 380 -QQQMIQQLLQEMSNNN---GGLQSQSLGGHNANGNISKNTMGFGGHTPSLSGGSANVPG 547
Query: 401 GS 402
+
Sbjct: 548 NN 553
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.121 0.328
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,728,490
Number of Sequences: 63676
Number of extensions: 291215
Number of successful extensions: 5090
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 3048
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3807
length of query: 570
length of database: 12,639,632
effective HSP length: 102
effective length of query: 468
effective length of database: 6,144,680
effective search space: 2875710240
effective search space used: 2875710240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144645.6


