

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144618.8 + phase: 0 /pseudo
(558 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
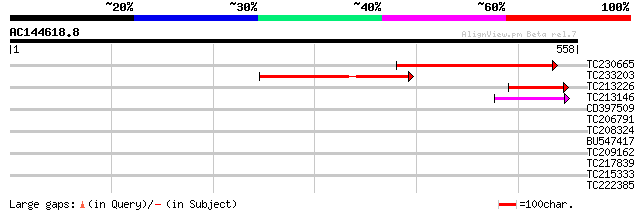
Score E
Sequences producing significant alignments: (bits) Value
TC230665 129 3e-30
TC233203 similar to UP|Q9FM34 (Q9FM34) Dbj|BAA95714.1, partial (... 120 2e-27
TC213226 104 1e-22
TC213146 54 2e-07
CD397509 homologue to SP|O23429|V724_ Vesicle-associated membran... 39 0.005
TC206791 similar to UP|Q8L4Y5 (Q8L4Y5) Actin-related protein 7, ... 31 1.3
TC208324 similar to UP|WRK6_ARATH (Q9C519) WRKY transcription fa... 30 2.2
BU547417 30 2.8
TC209162 similar to UP|Q9XHE5 (Q9XHE5) Microtubule-associated pr... 30 3.7
TC217839 similar to UP|Q9LM23 (Q9LM23) T10O22.24, partial (26%) 29 4.8
TC215333 similar to UP|P93344 (P93344) Aldehyde dehydrogenase (N... 29 6.3
TC222385 weakly similar to PIR|G86367|G86367 protein F28C11.3 [i... 28 8.3
>TC230665
Length = 914
Score = 129 bits (324), Expect = 3e-30
Identities = 72/159 (45%), Positives = 105/159 (65%)
Frame = +3
Query: 381 GRIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWSLLQCLLTSAKSNSDGLVISDDIELT 440
G ++ +G+I+ ++IEE + +SI LPA+ L+Q LL SAK NS G++ISD+ ELT
Sbjct: 15 GEKMVSEGIITREDIEEPKSGNGSRVISIGLPAYCLVQGLLRSAKLNSVGIMISDETELT 194
Query: 441 RMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVMRSKNDIPEDWDSTGFPSK 500
N P++ F+WF+ PLLI+KEQ+K L SEE L +LV+ + S G +
Sbjct: 195 ITNRPRETFFDWFVNPLLIIKEQIKAENLSASEEDYLCKLVLLGGDAERLKKSSVGPAPE 374
Query: 501 DNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKIL 539
+V+RA+L+A+ RRLQGI SMSR PTF+R+F +LV+ L
Sbjct: 375 SDVKRAELEALARRLQGITKSMSRFPTFKRRFNDLVRSL 491
>TC233203 similar to UP|Q9FM34 (Q9FM34) Dbj|BAA95714.1, partial (12%)
Length = 467
Score = 120 bits (300), Expect = 2e-27
Identities = 61/151 (40%), Positives = 92/151 (60%)
Frame = +3
Query: 247 IILWPLAVLGAVLAAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYL 306
I+LWPLAV+GAVLA+++ SF L Y G + +QE S G YIV+ VSL+DEY ND+L +
Sbjct: 3 IVLWPLAVVGAVLASVLASFVLGGYAGFITYQESSFLFGLGYIVAAVSLYDEYSNDILDM 182
Query: 307 REGSCLPRPIYRKNVKHAVERKKLEGIDHNLKNRRDSSQNSKHTLQQSRSMKWKIQQYKP 366
+EGSC PRP YRK + + R I +++ TL + S+K I ++KP
Sbjct: 183 KEGSCFPRPRYRKITELKLSRTPSRSIS------LPKTKSLTKTLSHAYSLKDNIPEFKP 344
Query: 367 VQVWDWFFKSCEVNGRIVLRDGLISVKEIEE 397
++ D FK C G ++ +G+I+ ++I+E
Sbjct: 345 FELLDGLFKECHQLGESLVSEGVITHEDIQE 437
>TC213226
Length = 438
Score = 104 bits (259), Expect = 1e-22
Identities = 51/59 (86%), Positives = 55/59 (92%)
Frame = +1
Query: 492 WDSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKILYIEALQASASA 550
WDSTGFPS D VRRAQLQAIIRRLQGI +SMSRMPTFRR+FRNL+K+LYIEALQA ASA
Sbjct: 10 WDSTGFPSDDTVRRAQLQAIIRRLQGIVASMSRMPTFRRRFRNLIKVLYIEALQAGASA 186
>TC213146
Length = 416
Score = 53.5 bits (127), Expect = 2e-07
Identities = 26/74 (35%), Positives = 43/74 (57%)
Frame = +3
Query: 478 KELVMRSKNDIPEDWDSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVK 537
K ++ S E WD+ G D +R AQ++ I RR+ G+ +S++PT++RKFR +VK
Sbjct: 24 KVVLFGSNKQRMEAWDNGGLMIHDALRAAQIEGISRRMIGMIRGISKLPTYKRKFRQIVK 203
Query: 538 ILYIEALQASASAK 551
L +L+ S +
Sbjct: 204 ALVTHSLEKDVSER 245
>CD397509 homologue to SP|O23429|V724_ Vesicle-associated membrane protein
724 (AtVAMP724) (SYBL1-like protein). [Mouse-ear cress],
partial (19%)
Length = 618
Score = 39.3 bits (90), Expect = 0.005
Identities = 17/32 (53%), Positives = 25/32 (78%)
Frame = -3
Query: 508 LQAIIRRLQGIGSSMSRMPTFRRKFRNLVKIL 539
+ +++ RLQGI SMSR PTF+R+F +LV+ L
Sbjct: 478 MSSLLFRLQGITKSMSRFPTFKRRFNDLVRSL 383
>TC206791 similar to UP|Q8L4Y5 (Q8L4Y5) Actin-related protein 7, partial
(63%)
Length = 1117
Score = 31.2 bits (69), Expect = 1.3
Identities = 15/51 (29%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Frame = +1
Query: 36 VGNSAVIIG-LWTAHVFWTYYCVARLYFLSKFFVICFICSNSISLINCWPL 85
+G +++G L+++HV+W +A L LS F +C + N +CW +
Sbjct: 310 LGEKDILLGKLYSSHVYWV*RLMALLISLSVLFQLCHLIING----SCWKI 450
>TC208324 similar to UP|WRK6_ARATH (Q9C519) WRKY transcription factor 6 (WRKY
DNA-binding protein 6) (AtWRKY6), partial (19%)
Length = 1638
Score = 30.4 bits (67), Expect = 2.2
Identities = 25/95 (26%), Positives = 45/95 (47%), Gaps = 3/95 (3%)
Frame = +2
Query: 324 AVERKKLEGIDHNLKN-RRDSSQNS--KHTLQQSRSMKWKIQQYKPVQVWDWFFKSCEVN 380
A+++KKL N ++ ++DS QN+ K L R F + ++N
Sbjct: 440 AMQQKKLSSSPRNNEDMQKDSQQNNMEKPALPSCRQ----------------FLNTGKIN 571
Query: 381 GRIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWS 415
R++L++ I +EE F+ +CKK + + A S
Sbjct: 572 NRVILQEAKI----VEEQAFEASCKKARVSVRARS 664
>BU547417
Length = 643
Score = 30.0 bits (66), Expect = 2.8
Identities = 12/31 (38%), Positives = 17/31 (54%)
Frame = +2
Query: 49 HVFWTYYCVARLYFLSKFFVICFICSNSISL 79
H FW + + FLS F +C +C +S SL
Sbjct: 482 HTFWYRFSLNFCVFLSLFLFLCVLCFDSFSL 574
>TC209162 similar to UP|Q9XHE5 (Q9XHE5) Microtubule-associated protein,
partial (8%)
Length = 713
Score = 29.6 bits (65), Expect = 3.7
Identities = 15/53 (28%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Frame = -3
Query: 324 AVERKKLEGIDH-NLKNRRDSSQNSKHTLQQSRSMKWKIQQYKPVQVWDWFFK 375
++E ++LEG+DH + + QN + S W+ + + V WDW+ K
Sbjct: 174 SLEDQRLEGVDHWDR*LKFHCIQNFLYCTVMSEKCSWEDWRLEGVDHWDWYVK 16
>TC217839 similar to UP|Q9LM23 (Q9LM23) T10O22.24, partial (26%)
Length = 1532
Score = 29.3 bits (64), Expect = 4.8
Identities = 16/48 (33%), Positives = 26/48 (53%), Gaps = 2/48 (4%)
Frame = -2
Query: 37 GNSAVIIGLWTA--HVFWTYYCVARLYFLSKFFVICFICSNSISLINC 82
GNS+++ G W + ++ +R L+ F +I CSNSIS + C
Sbjct: 709 GNSSLLQGTWMDL*YSRTLFFLCSRYNSLNSFQIIYIKCSNSISTLFC 566
>TC215333 similar to UP|P93344 (P93344) Aldehyde dehydrogenase (NAD+) ,
partial (73%)
Length = 1405
Score = 28.9 bits (63), Expect = 6.3
Identities = 18/51 (35%), Positives = 25/51 (48%), Gaps = 2/51 (3%)
Frame = +3
Query: 38 NSAVIIGLWTAHVFWTYYCVARLYFLSKFFVICFICSNSIS--LINCWPLK 86
+S V + WT W ++C + VICFICS +IS +CW K
Sbjct: 198 SSHVCLEGWTGIGLWQHHCSQNS*ADT---VICFICSKTIS*GWTSCWCFK 341
>TC222385 weakly similar to PIR|G86367|G86367 protein F28C11.3 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(9%)
Length = 364
Score = 28.5 bits (62), Expect = 8.3
Identities = 14/42 (33%), Positives = 21/42 (49%)
Frame = -2
Query: 413 AWSLLQCLLTSAKSNSDGLVISDDIELTRMNGPKDRVFEWFI 454
AW L+C + K+NSDG+ + +E G + FE I
Sbjct: 360 AWCQLRCSASREKANSDGVSV*KSVECEDRTGEV*QYFELLI 235
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.333 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,511,284
Number of Sequences: 63676
Number of extensions: 439019
Number of successful extensions: 3081
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 3051
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3078
length of query: 558
length of database: 12,639,632
effective HSP length: 102
effective length of query: 456
effective length of database: 6,144,680
effective search space: 2801974080
effective search space used: 2801974080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144618.8


