

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144564.13 - phase: 0
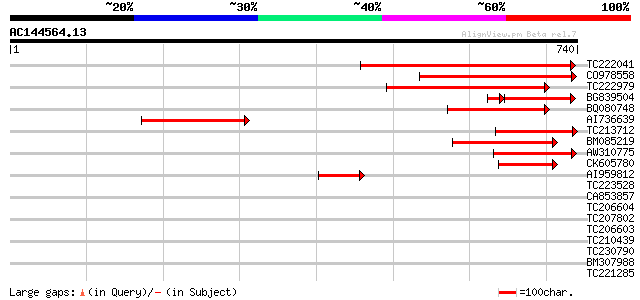
(740 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222041 similar to UP|Q8LBQ6 (Q8LBQ6) Isp4-like protein, partia... 549 e-156
CO978558 289 4e-78
TC222979 similar to UP|Q8H0V0 (Q8H0V0) Isp4 like protein, partia... 280 2e-75
BG839504 similar to GP|20466754|gb Isp4-like protein {Arabidopsi... 176 3e-52
BQ080748 weakly similar to PIR|T08925|T08 isp4 protein homolog T... 173 2e-43
AI736639 171 8e-43
TC213712 similar to UP|Q9LWW1 (Q9LWW1) ESTs D25093(R3139) (Gluta... 161 1e-39
BM085219 weakly similar to GP|9758213|dbj sexual differentiation... 159 3e-39
AW310775 weakly similar to PIR|T08925|T08 isp4 protein homolog T... 135 5e-32
CK605780 87 3e-17
AI959812 77 3e-14
TC223528 weakly similar to UP|Q8H0V0 (Q8H0V0) Isp4 like protein,... 41 0.002
CA853857 similar to PIR|H96681|H96 protein F1E22.10 [imported] -... 39 0.011
TC206604 similar to UP|Q9LSD4 (Q9LSD4) Gb|AAF23830.1, partial (37%) 38 0.014
TC207802 similar to UP|Q6R3L0 (Q6R3L0) Metal-nicotianamine trans... 35 0.092
TC206603 similar to UP|Q9LSD4 (Q9LSD4) Gb|AAF23830.1, partial (47%) 35 0.16
TC210439 similar to GB|BAB02316.1|11994357|AB017071 delta 9 desa... 33 0.60
TC230790 similar to UP|Q9FJC8 (Q9FJC8) EspB-like protein, partia... 32 1.3
BM307988 31 1.7
TC221285 similar to UP|Q6R3L0 (Q6R3L0) Metal-nicotianamine trans... 31 2.3
>TC222041 similar to UP|Q8LBQ6 (Q8LBQ6) Isp4-like protein, partial (38%)
Length = 1012
Score = 549 bits (1414), Expect = e-156
Identities = 252/281 (89%), Positives = 268/281 (94%)
Frame = +2
Query: 458 LAFIFTLPISIITATTNQTPGLNIITEYIFGIIYPGRPIANVCFKTYGYISMAQAVSFLS 517
LAF FTLPISIITATTNQTPGLNIITEY+FG+IYPGRPIANVCFKTYGYISMAQAVSFLS
Sbjct: 2 LAFGFTLPISIITATTNQTPGLNIITEYVFGLIYPGRPIANVCFKTYGYISMAQAVSFLS 181
Query: 518 DFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPKGSPWTCPS 577
DFKLGHYMKIPPRSMFLVQFIGT+LAGTINIGVAWWLL+S+KNIC+ DLLP+GSPWTCP
Sbjct: 182 DFKLGHYMKIPPRSMFLVQFIGTILAGTINIGVAWWLLNSIKNICHDDLLPEGSPWTCPG 361
Query: 578 DRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHKAFPKQSWIPLIN 637
DRVFFDASV+WGLVGPKRIFGS G YS +NWFFLGGA+GPI+VWLLHKAFPKQSWIPLIN
Sbjct: 362 DRVFFDASVIWGLVGPKRIFGSQGNYSAMNWFFLGGALGPIIVWLLHKAFPKQSWIPLIN 541
Query: 638 LPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDAGVAFMA 697
LPVLLGATGMMPPAT LNYN+WI VGTIFNFFIFRYRKKWWQRYNYVLSAALD+GVAFM
Sbjct: 542 LPVLLGATGMMPPATPLNYNAWIFVGTIFNFFIFRYRKKWWQRYNYVLSAALDSGVAFMT 721
Query: 698 VLLYLALGLENVSLNWWGTAGEHCPLAACPTAKGIVVDGCP 738
V+LY +LGLEN SLNWWG GEHCPLAACPTAKGI+VDGCP
Sbjct: 722 VMLYFSLGLENKSLNWWGNDGEHCPLAACPTAKGIIVDGCP 844
>CO978558
Length = 721
Score = 289 bits (739), Expect = 4e-78
Identities = 127/204 (62%), Positives = 157/204 (76%)
Frame = -2
Query: 536 QFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKR 595
Q +GT+L+ I AWWL+ ++ ++C+ +L SPWTCP D VFFDASV+WGL+GP+R
Sbjct: 717 QVVGTILSVFIYTVTAWWLMRTIPHLCDTTMLDPDSPWTCPMDNVFFDASVIWGLLGPRR 538
Query: 596 IFGSLGEYSTLNWFFLGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALN 655
IFG LGEY+ +N FFLGGAI P LVWL HKAFP+Q WI LI++PVLLGAT MMPPATA+N
Sbjct: 537 IFGDLGEYAKVNLFFLGGAIAPFLVWLAHKAFPEQRWITLIHMPVLLGATSMMPPATAVN 358
Query: 656 YNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLNWWG 715
+ SWI+VG + + +RYR++WW+R NYVLS LDAG AFM +LL+LAL N+ LNWWG
Sbjct: 357 FTSWILVGFLSGYIAYRYRQEWWKRCNYVLSGGLDAGTAFMTILLFLALNNNNIMLNWWG 178
Query: 716 TAGEHCPLAACPTAKGIVVDGCPV 739
E CPLA CPT KGIVV GCPV
Sbjct: 177 NNPEGCPLANCPTEKGIVVKGCPV 106
>TC222979 similar to UP|Q8H0V0 (Q8H0V0) Isp4 like protein, partial (30%)
Length = 661
Score = 280 bits (715), Expect = 2e-75
Identities = 128/212 (60%), Positives = 155/212 (72%)
Frame = +2
Query: 493 GRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAW 552
G+PIAN+ FK YG IS A+SFLSD KLGHYMKIPPR M+ Q GT+ AG N+ VAW
Sbjct: 2 GKPIANLLFKIYGRISTVHALSFLSDLKLGHYMKIPPRCMYTAQLEGTLFAGVENLAVAW 181
Query: 553 WLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLG 612
W+ DS+K+IC D L SPWTCP RV FDASV+WGL+GPKR+FG G Y L W FL
Sbjct: 182 WM*DSIKDICMDDKLHHDSPWTCPKYRVTFDASVIWGLIGPKRLFGPGGLYRNLVWLFLI 361
Query: 613 GAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFR 672
GA P+ +W+L K FP++ WIPLIN+PV+ MPPAT N SW++ G IFN+F+FR
Sbjct: 362 GAELPVPIWVLSKIFPEKKWIPLINIPVITYRFAGMPPATPANIASWLVTGMIFNYFVFR 541
Query: 673 YRKKWWQRYNYVLSAALDAGVAFMAVLLYLAL 704
Y K+WWQ+YNYVLSAALDA AFM VL++ AL
Sbjct: 542 YNKRWWQKYNYVLSAALDAXTAFMGVLIFFAL 637
>BG839504 similar to GP|20466754|gb Isp4-like protein {Arabidopsis thaliana},
partial (15%)
Length = 499
Score = 176 bits (446), Expect(2) = 3e-52
Identities = 77/93 (82%), Positives = 81/93 (86%)
Frame = +3
Query: 646 GMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALG 705
GMMPPAT LNYN+WI VGTIFNFFIFRYR KWW RYNYVLSAALD+GVAFM VL Y ALG
Sbjct: 69 GMMPPATPLNYNAWIFVGTIFNFFIFRYRXKWWXRYNYVLSAALDSGVAFMTVLXYFALG 248
Query: 706 LENVSLNWWGTAGEHCPLAACPTAKGIVVDGCP 738
LEN S NWWG GEHCPLA CPTAKG++VDGCP
Sbjct: 249 LENKSXNWWGNDGEHCPLAVCPTAKGVIVDGCP 347
Score = 48.5 bits (114), Expect(2) = 3e-52
Identities = 21/23 (91%), Positives = 21/23 (91%)
Frame = +2
Query: 624 HKAFPKQSWIPLINLPVLLGATG 646
HKAFPKQSWIP INLPV LGATG
Sbjct: 2 HKAFPKQSWIPXINLPVXLGATG 70
>BQ080748 weakly similar to PIR|T08925|T08 isp4 protein homolog T15N24.40 -
Arabidopsis thaliana, partial (17%)
Length = 421
Score = 173 bits (439), Expect = 2e-43
Identities = 74/133 (55%), Positives = 95/133 (70%)
Frame = +3
Query: 572 PWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHKAFPKQS 631
PWTCP D VF++AS++WG+VGPKR+F G Y +NWFFL G + P+ VWLL K FP
Sbjct: 3 PWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPGMNWFFLIGLLAPLPVWLLSKKFPNHK 182
Query: 632 WIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDA 691
WI LIN P+++ +PP ++NY +W IVG FNF+++R K WW R+ Y+LSAALDA
Sbjct: 183 WIQLINFPIIIAGASNIPPFRSVNYITWGIVGIFFNFYVYRKFKAWWARHTYILSAALDA 362
Query: 692 GVAFMAVLLYLAL 704
GVAFM V LY AL
Sbjct: 363 GVAFMGVALYFAL 401
>AI736639
Length = 427
Score = 171 bits (434), Expect = 8e-43
Identities = 76/141 (53%), Positives = 100/141 (70%), Gaps = 1/141 (0%)
Frame = +2
Query: 173 GWAGLLRKYVVEPAHMWWPGTLVQVSLFRTLHEKDDNPHQ-FSRAKFFFIALVCSFTWYI 231
GWAGL RK++VEP MWWP LVQVSLF LHEK P +R +FF + LV +Y+
Sbjct: 5 GWAGLFRKFLVEPGEMWWPSNLVQVSLFSALHEKSKRPKGGTTRTQFFLLVLVSGLAYYV 184
Query: 232 VPGYLFTTLTSISWVCWVFSKSVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFF 291
PGYLF+ LTS SW+CW+ KSV QQ+GSG+ GLG+ A +DWS ++++L SPL SP+F
Sbjct: 185 FPGYLFSMLTSFSWMCWLAPKSVLVQQLGSGLRGLGIAAFGIDWSTISAYLGSPLASPWF 364
Query: 292 AIVNVFVGYALLVYAVIPIAY 312
A N+ VG+ L++Y + PIAY
Sbjct: 365 ATANIAVGFFLVMYVMTPIAY 427
>TC213712 similar to UP|Q9LWW1 (Q9LWW1) ESTs D25093(R3139) (Glutathione
transporter), partial (14%)
Length = 393
Score = 161 bits (407), Expect = 1e-39
Identities = 72/106 (67%), Positives = 86/106 (80%)
Frame = +2
Query: 635 LINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDAGVA 694
LIN+PVL+GATGMMPPATA+NY SWII G + F +FRY+ +WW+R+NYVLS ALDAG+A
Sbjct: 11 LINMPVLIGATGMMPPATAVNYTSWIIAGFLSGFVVFRYKPEWWKRHNYVLSGALDAGLA 190
Query: 695 FMAVLLYLALGLENVSLNWWGTAGEHCPLAACPTAKGIVVDGCPVF 740
FM VLLYL LGLE++SLNWWG + C LA CPTAK + V GC VF
Sbjct: 191 FMGVLLYLCLGLEDISLNWWGNDLDGCTLAHCPTAKVVEVQGCAVF 328
>BM085219 weakly similar to GP|9758213|dbj sexual differentiation process
protein ISP4-like {Arabidopsis thaliana}, partial (17%)
Length = 426
Score = 159 bits (403), Expect = 3e-39
Identities = 71/139 (51%), Positives = 96/139 (68%), Gaps = 1/139 (0%)
Frame = -2
Query: 578 DRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHKAFPKQSWIPLIN 637
D VF++AS++WG+VGP+R+F G Y +NWFFL G + P+ VWLL + FP WI LIN
Sbjct: 425 DEVFYNASIIWGVVGPQRMFTKDGIYPGMNWFFLIGLLAPVPVWLLARKFPNHKWIELIN 246
Query: 638 LPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDAGVAFMA 697
+P+++ G +PPA ++NY +W VG FNF+++ K WW R+ Y+LSAALDAGVAFM
Sbjct: 245 MPLIIAGGGGIPPARSVNYITWGFVGIFFNFYVYSKFKAWWARHTYILSAALDAGVAFMG 66
Query: 698 VLLYLALGLENV-SLNWWG 715
V+LY AL V WWG
Sbjct: 65 VILYFALQNYGVFGPIWWG 9
>AW310775 weakly similar to PIR|T08925|T08 isp4 protein homolog T15N24.40 -
Arabidopsis thaliana, partial (14%)
Length = 466
Score = 135 bits (341), Expect = 5e-32
Identities = 60/110 (54%), Positives = 81/110 (73%), Gaps = 2/110 (1%)
Frame = -1
Query: 632 WIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDA 691
WI LIN+P+++ +G +PPA ++NY +W +VG +FNF+++ K WW R+NY+LSAALDA
Sbjct: 457 WIELINMPLIIAGSGGIPPARSVNYITWGMVGILFNFYVYNKFKAWWARHNYILSAALDA 278
Query: 692 GVAFMAVLLYLALGLENV-SLNWWG-TAGEHCPLAACPTAKGIVVDGCPV 739
GVAFM V+LY AL ++ WWG A +HCPLA CPTA GI+ GCPV
Sbjct: 277 GVAFMGVILYFALQYYDIFGPTWWGQDADDHCPLAKCPTAPGIISHGCPV 128
>CK605780
Length = 491
Score = 87.0 bits (214), Expect = 3e-17
Identities = 42/78 (53%), Positives = 48/78 (60%), Gaps = 1/78 (1%)
Frame = -3
Query: 639 PVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDAGVAFMAV 698
P + GA G +PPAT W+IVG IFN+FI R WW RYNYVLS ALD G A V
Sbjct: 471 PSIFGAAGQIPPATCWYLGQWVIVGLIFNYFIRRAYFGWWTRYNYVLSGALDIGTALCIV 292
Query: 699 LLYLALGLENVSL-NWWG 715
+ LALGL S +WWG
Sbjct: 291 VSALALGLSGASFPDWWG 238
>AI959812
Length = 193
Score = 77.0 bits (188), Expect = 3e-14
Identities = 27/60 (45%), Positives = 43/60 (71%)
Frame = -2
Query: 404 KEDIHTKLMKNYKDIPSWWFYLLLGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFIFT 463
K DIHT+LM+ YK +P+WWFY++L + + IC + N+ +Q+PWWG+L+A A++ T
Sbjct: 180 KIDIHTRLMRRYKSVPTWWFYIILAANIALIIFICEYYNESLQLPWWGVLLACAISIFRT 1
>TC223528 weakly similar to UP|Q8H0V0 (Q8H0V0) Isp4 like protein, partial
(4%)
Length = 399
Score = 40.8 bits (94), Expect = 0.002
Identities = 14/27 (51%), Positives = 17/27 (62%)
Frame = +3
Query: 713 WWGTAGEHCPLAACPTAKGIVVDGCPV 739
WWG +HC LA CPTA + GCP+
Sbjct: 24 WWGLDADHCHLAKCPTAPDVHAKGCPL 104
>CA853857 similar to PIR|H96681|H96 protein F1E22.10 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 531
Score = 38.5 bits (88), Expect = 0.011
Identities = 36/113 (31%), Positives = 54/113 (46%), Gaps = 1/113 (0%)
Frame = -2
Query: 509 MAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLP 568
++ A DFK G+ PRSMFL Q +GT + I+ V W + N+ +P
Sbjct: 353 VSXASDLTQDFKTGYMTLASPRSMFLSQVLGTAMGCVISPCVFWLFYKAFGNLG----IP 186
Query: 569 KGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYS-TLNWFFLGGAIGPILV 620
GS + P V+ + +++ G+ G F +L +Y TL F AIG LV
Sbjct: 185 -GSAYPAPYALVYRNMAIL-GVDG----FSALPQYCLTLCCVFFVAAIGINLV 45
>TC206604 similar to UP|Q9LSD4 (Q9LSD4) Gb|AAF23830.1, partial (37%)
Length = 1091
Score = 38.1 bits (87), Expect = 0.014
Identities = 60/251 (23%), Positives = 102/251 (39%), Gaps = 6/251 (2%)
Frame = +2
Query: 448 PWWGLLIASALAFIFTLPISIIT---ATTNQTPGLNIITEYIFGIIYPGRPIANVCFKTY 504
P ++A ALAF + + A+T GL II + G IA V
Sbjct: 8 PQGSYILAPALAFCNSYGTGLTDWSLASTYGKIGLFIIAA---AVGQNGGVIAGVASCAV 178
Query: 505 GYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNK 564
+A A + DFK G+ +SMF+ Q IGT + G + + +W+ + +I +
Sbjct: 179 MMSIVATAADLMQDFKTGYLTLSSAKSMFVSQLIGTAM-GCVIAPLTFWMFWTAFDIGSP 355
Query: 565 DLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEY--STLNWFFLGGAIGPILVWL 622
D P+ P +F + +++ G+ G F L +Y FF +L +
Sbjct: 356 D-----GPYKAPYAVIFREMAIL-GVQG----FSELPKYCLEMCGGFFAAALAINLLRDV 505
Query: 623 LHKAFPKQSWIPL-INLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRY 681
K F + IP+ + +P +GA + + VGT+ F R +K + Y
Sbjct: 506 TPKKFSQYIPIPMAMAVPFYIGAYFAVD----------MFVGTVILFVWERLNRKDAEDY 655
Query: 682 NYVLSAALDAG 692
+++ L G
Sbjct: 656 AGAVASGLICG 688
>TC207802 similar to UP|Q6R3L0 (Q6R3L0) Metal-nicotianamine transporter YSL1,
partial (29%)
Length = 1137
Score = 35.4 bits (80), Expect = 0.092
Identities = 40/148 (27%), Positives = 73/148 (49%), Gaps = 8/148 (5%)
Frame = +1
Query: 505 GYISMAQAVS--FLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNIC 562
G I +VS + DFK HY + PR+MF+ Q IG + G + ++++L ++
Sbjct: 13 GLIKSVISVSCILMQDFKTAHYTRTSPRAMFICQVIG-IAMGCVTAPLSFFLYYKAFDVG 189
Query: 563 NKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYS-TLNWFFLGGAIGPILVW 621
N P G + P ++ + +++ G+ G F +L ++ L + F AIG V
Sbjct: 190 N----PHGE-FKAPYALIYRNMAII-GVQG----FSALPQHCLQLCFGFFAFAIG---VN 330
Query: 622 LLHKAFPKQ--SWIPL---INLPVLLGA 644
++ P++ W+PL + +P L+GA
Sbjct: 331 MIRDFAPQKIGKWMPLPMVMAVPFLVGA 414
>TC206603 similar to UP|Q9LSD4 (Q9LSD4) Gb|AAF23830.1, partial (47%)
Length = 1062
Score = 34.7 bits (78), Expect = 0.16
Identities = 42/176 (23%), Positives = 76/176 (42%), Gaps = 6/176 (3%)
Frame = +1
Query: 418 IPSWWF---YLLLGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIIT---A 471
IP+W+ Y+ L + ++ I IF + + ++A ALAF + + A
Sbjct: 424 IPTWFAASGYVGLAAISIATIPI-IFPPLKWYLVLCSYILAPALAFCNSYGTGLTDWSLA 600
Query: 472 TTNQTPGLNIITEYIFGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRS 531
+T GL II + G IA V +A A + DFK G+ +S
Sbjct: 601 STYGKIGLFIIAA---AVGQNGGVIAGVAACAVMMSIVATAADLMQDFKTGYLTLSSAKS 771
Query: 532 MFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVV 587
MF+ Q IGT + G + + +W+ + ++ + D P+ P +F + +++
Sbjct: 772 MFVSQLIGTAM-GCVIAPLTFWMFWTAFDVGSPD-----GPYKAPYAVIFREMAIL 921
>TC210439 similar to GB|BAB02316.1|11994357|AB017071 delta 9 desaturase-like
protein {Arabidopsis thaliana;} , partial (40%)
Length = 820
Score = 32.7 bits (73), Expect = 0.60
Identities = 31/115 (26%), Positives = 49/115 (41%), Gaps = 19/115 (16%)
Frame = +3
Query: 105 SKRFSFNP---------GPFNMKEHVLITIFANAGAAFGSG------SSYAVGIVNIIKA 149
+K+ +FNP PF ++ +T ANA AAFG G S + +++ ++
Sbjct: 156 TKKLNFNPTQTKISLQNNPFTY--NLAVTAKANANAAFGEGFKEEGASRHRKILLSEVEV 329
Query: 150 FYGRNISFLAAWLLIITTQVLGYGWAGLLRKY----VVEPAHMWWPGTLVQVSLF 200
R + F W L +G AG++ V P H WP V V+L+
Sbjct: 330 KRERRVFFGRKW------NSLDFGTAGIVLAMHLLCVFAPFHFNWPAFRVAVALY 476
>TC230790 similar to UP|Q9FJC8 (Q9FJC8) EspB-like protein, partial (32%)
Length = 861
Score = 31.6 bits (70), Expect = 1.3
Identities = 18/50 (36%), Positives = 26/50 (52%)
Frame = +2
Query: 516 LSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKD 565
+ DFK GH PRSM L Q IGT + G + + ++L ++ N D
Sbjct: 92 MHDFKTGHLTFTSPRSMLLSQAIGTAI-GCVVAPLTFFLFYKAFDVGNPD 238
>BM307988
Length = 455
Score = 31.2 bits (69), Expect = 1.7
Identities = 16/48 (33%), Positives = 22/48 (45%)
Frame = +1
Query: 419 PSWWFYLLLGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPI 466
PS WF L F+ S+ IFL + + W L+ + FTL I
Sbjct: 295 PSSWFVLFCSFFFIFSVACLIFLEQNLNIILWEFLVLFSENLSFTLLI 438
>TC221285 similar to UP|Q6R3L0 (Q6R3L0) Metal-nicotianamine transporter YSL1,
partial (32%)
Length = 945
Score = 30.8 bits (68), Expect = 2.3
Identities = 13/35 (37%), Positives = 19/35 (54%)
Frame = +1
Query: 512 AVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTI 546
A + DFK +Y P++MF+ Q +GT L I
Sbjct: 85 ACXLMLDFKTAYYTCTSPKAMFICQLVGTALGCVI 189
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,652,904
Number of Sequences: 63676
Number of extensions: 635715
Number of successful extensions: 5131
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 4859
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5039
length of query: 740
length of database: 12,639,632
effective HSP length: 104
effective length of query: 636
effective length of database: 6,017,328
effective search space: 3827020608
effective search space used: 3827020608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144564.13


