

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144563.2 + phase: 0
(576 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
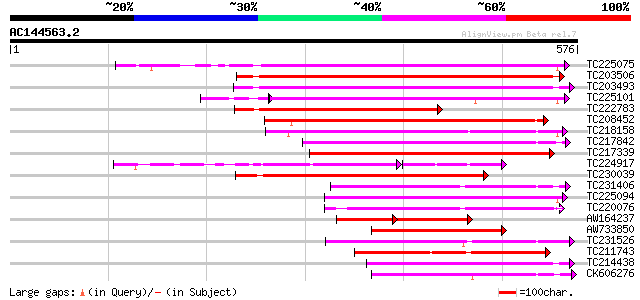
Score E
Sequences producing significant alignments: (bits) Value
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 297 8e-81
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 285 4e-77
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 283 1e-76
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 255 3e-71
TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylester... 261 5e-70
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 250 1e-66
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 249 3e-66
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 238 4e-63
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 234 6e-62
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 168 8e-61
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 229 2e-60
TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (... 213 2e-55
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 210 2e-54
TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 pr... 207 1e-53
AW164237 122 1e-53
AW733850 205 5e-53
TC231526 similar to UP|PME_MEDSA (Q42920) Pectinesterase precurs... 192 4e-49
TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 191 6e-49
TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (F... 190 1e-48
CK606276 177 9e-45
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 297 bits (761), Expect = 8e-81
Identities = 172/468 (36%), Positives = 252/468 (53%), Gaps = 7/468 (1%)
Frame = +1
Query: 108 ALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQI----ADMRNWLSAVISYRQACMEGF 163
A+ DC DL+ + D L + N + H+ +D+R WLSA +++ + CMEGF
Sbjct: 250 AIADCLDLLDLSSDVLSWALSA--SQNPKGKHNSTGNLSSDLRTWLSAALAHPETCMEGF 423
Query: 164 DDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSE 223
+ N K + V+A +V+ + +L Q V PA + +
Sbjct: 424 EGTNSIVKGL--------------VSAGIGQVVSLVEQLLAQ------VLPAQDQF---D 534
Query: 224 VTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGN 283
+PSWI +RKLL + + P+ VA DGSG + I A+ + P +
Sbjct: 535 AASSKGQFPSWIKPKERKLLQAIA-------VTPDVTVALDGSGNYAKIMDAVLAAPDYS 693
Query: 284 KGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTA 343
R+VI VK GVY E + + K NI++ G G T+++G +S G T ++ATFA +
Sbjct: 694 MKRFVILVKKGVYVENVEIKKKKWNIMILGQGMDATVISGNRSVVDGWTTFRSATFAVSG 873
Query: 344 MGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVS 403
GFI + ++F+NTAGP+ HQAVA R+ D+S C I GYQDSLY + RQ++R+C +S
Sbjct: 874 RGFIARDISFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDCTIS 1053
Query: 404 GTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALF 463
GTVD+IFG + + Q+ + V+K Q NTITA G N TG Q CNI ++ L
Sbjct: 1054GTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHGRKDPNEPTGFSFQFCNITADSDLI 1233
Query: 464 PERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGA 523
P T ++YLGRPWK ++TV M+S + + I +GW W G +T YYAEY NTG GA
Sbjct: 1234PSVGTAQTYLGRPWKSYSRTVFMQSYMSEVIGAEGWLEWNGNFALDTLYYAEYMNTGAGA 1413
Query: 524 NVARRVKWKGYHGVISRAEANKFTAGIWLQAG---PKSAAEWLNGLHV 568
VA RVKW GYH + ++A+ FT +++ P + + GL V
Sbjct: 1414GVANRVKWPGYHALNDSSQASNFTVSQFIEGNLWLPSTGVTFTAGLTV 1557
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 285 bits (729), Expect = 4e-77
Identities = 146/334 (43%), Positives = 204/334 (60%), Gaps = 1/334 (0%)
Frame = +2
Query: 231 YPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIY 290
+P W++ DR+LL+ + I + V+K G+G KTI A+ P+ + R++IY
Sbjct: 11 FPGWLNKRDRRLLSLPL-----SAIQADITVSKTGNGTVKTIAEAIKKAPEHSTRRFIIY 175
Query: 291 VKAGVYDEY-ITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGK 349
V+AG Y+E + V + N++ GDG +T++TG+K+ G+ T +A+FA + GFI +
Sbjct: 176 VRAGRYEENNLKVGRKKTNVMFIGDGKGKTVITGKKNVIDGMTTFHSASFAASGAGFIAR 355
Query: 350 AMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFI 409
+TFEN AGP HQAVA R D + + C+IVGYQDS YV SNRQ++R C + GTVDFI
Sbjct: 356 DITFENYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFI 535
Query: 410 FGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTI 469
FG++A + Q I RKP Q NTITA N NTGI + +C I+P L P + +
Sbjct: 536 FGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISLHNCRILPAPDLAPVKGSF 715
Query: 470 RSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRV 529
+YLGRPWK ++TV + S +GD IHP GW W G+ NT YY EY N GPGA V +RV
Sbjct: 716 PTYLGRPWKQYSRTVYLVSYMGDHIHPRGWLEWNGDFALNTLYYGEYMNYGPGAAVGQRV 895
Query: 530 KWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWL 563
+W GY + S EAN+FT ++ S + WL
Sbjct: 896 QWPGYRVIKSTMEANRFTVAQFI-----SGSAWL 982
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 283 bits (724), Expect = 1e-76
Identities = 146/348 (41%), Positives = 208/348 (58%), Gaps = 2/348 (0%)
Frame = +1
Query: 228 DQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRY 287
+ +P+W++ DR+LL+ + I + VV+KDG+G KTI A+ P+ + R
Sbjct: 130 EDNFPTWLNGRDRRLLSLPL-----SQIQADIVVSKDGNGTVKTIAEAIKKVPEYSSRRI 294
Query: 288 VIYVKAGVYDE-YITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGF 346
+IY++AG Y+E + + + N++ GDG +T++TG +++ + T TA+FA + GF
Sbjct: 295 IIYIRAGRYEEDNLKLGRKKTNVMFIGDGKGKTVITGGRNYYQNLTTFHTASFAASGSGF 474
Query: 347 IGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTV 406
I K MTFEN AGP HQAVA R D + + C+I+GYQD++YV SNRQ+YR C + GTV
Sbjct: 475 IAKDMTFENYAGPGRHQAVALRVGADHAVVYRCNIIGYQDTMYVHSNRQFYRECDIYGTV 654
Query: 407 DFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPER 466
DFIFG++A + Q+ T+ RKP Q NTITA N NTGI I +C I+ L +
Sbjct: 655 DFIFGNAAVVFQNCTLWARKPMAQQKNTITAQNRKDPNQNTGISIHNCRIMATPDLEASK 834
Query: 467 FTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQ-NHNTCYYAEYANTGPGANV 525
+ +YLGRPWK A+TV M S IGD +HP GW W +TCYY EY N GPG+ +
Sbjct: 835 GSYPTYLGRPWKLYARTVFMLSYIGDHVHPRGWLEWNTSSFALDTCYYGEYMNYGPGSAL 1014
Query: 526 ARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
+RV W GY + S EA++FT G ++ S + WL V G
Sbjct: 1015GQRVNWAGYRAINSTVEASRFTVGQFI-----SGSSWLPSTGVAFIAG 1143
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 255 bits (651), Expect(2) = 3e-71
Identities = 133/310 (42%), Positives = 184/310 (58%), Gaps = 6/310 (1%)
Frame = +3
Query: 265 GSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGR 324
G + + A+ + P + RYVI++K GVY E + + K N++M GDG TI++G
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGDGMDATIISGN 386
Query: 325 KSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGY 384
+SF G T ++ATFA + GFI + +TF+NTAGP+ HQAVA R+ D+S C I GY
Sbjct: 387 RSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGY 566
Query: 385 QDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMN 444
QDSLY + RQ+YR C +SGTVDFIFG + + Q+ I +K Q NTITA G +
Sbjct: 567 QDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAHGRKNPD 746
Query: 445 LNTGIVIQDCNIIPEAALFPERFTIRS---YLGRPWKYLAKTVVMESTIGDFIHPDGWTI 501
TG IQ CNI + L + S YLGRPWK ++T+ M+S I D + P+GW
Sbjct: 747 EPTGFSIQFCNISADYDLVNSVNSCNSTHTYLGRPWKPYSRTIFMQSYISDVLRPEGWLE 926
Query: 502 WQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAG---PKS 558
W G+ +T YYAEY N GPGA VA RVKW+GYH + ++A+ FT +++ P +
Sbjct: 927 WNGDFALDTLYYAEYMNYGPGAGVANRVKWQGYHVMNDSSQASNFTVSQFIEGNLCLPST 1106
Query: 559 AAEWLNGLHV 568
+ GL V
Sbjct: 1107GVTFTAGLGV 1136
Score = 32.7 bits (73), Expect(2) = 3e-71
Identities = 26/77 (33%), Positives = 39/77 (49%), Gaps = 3/77 (3%)
Frame = +2
Query: 195 IVTGLSDILQQFN-LKFDVKPASRRLLNSEVTVDDQG-YPSWISSSDRKLLAKMQRKNWR 252
+ TG+ ++ L VKP S S QG YPSW+ + +RKLL +
Sbjct: 23 VSTGIGQVMSLLQQLLTQVKPVSDHFSFSS----PQGQYPSWVKTGERKLL--------Q 166
Query: 253 ANIMP-NAVVAKDGSGQ 268
AN++ +AVV DG+G+
Sbjct: 167 ANVVSFDAVVTADGTGK 217
>TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylesterase
precursor , partial (29%)
Length = 630
Score = 261 bits (668), Expect = 5e-70
Identities = 124/211 (58%), Positives = 161/211 (75%)
Frame = +3
Query: 229 QGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYV 288
+GYP+W+S++DRKLLA++ ++P+A VAKDGSGQF T+ A+ SYPK ++GRY+
Sbjct: 9 EGYPTWVSAADRKLLAQLND----GAVLPHATVAKDGSGQFTTVLDAINSYPKKHQGRYI 176
Query: 289 IYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIG 348
IYVKAG+YDEYITV K N+L+YGDGP +TI+TGRK+F G KTM+TATF+ A F+
Sbjct: 177 IYVKAGIYDEYITVDKKKPNLLIYGDGPTKTIITGRKNFHEGTKTMRTATFSTVAEDFMA 356
Query: 349 KAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDF 408
K++ FENTAG +GHQAVA R QGD S C + GYQD+LY ++RQ+YRNC +SGT+DF
Sbjct: 357 KSIAFENTAGAEGHQAVALRVQGDRSVFFDCAMRGYQDTLYAHAHRQFYRNCEISGTIDF 536
Query: 409 IFGSSATLIQHSTIIVRKPGKGQFNTITADG 439
IFG S TLIQ+S I+VRKP Q N + ADG
Sbjct: 537 IFGYSTTLIQNSKILVRKPMPNQQNIVVADG 629
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 250 bits (638), Expect = 1e-66
Identities = 127/291 (43%), Positives = 177/291 (60%), Gaps = 3/291 (1%)
Frame = +1
Query: 260 VVAKDGSGQFKTIQAALASYPKGNK---GRYVIYVKAGVYDEYITVPKDAVNILMYGDGP 316
+V+ G + +I A+A+ P K G +++YV+ G+Y+EY+ +PK+ NIL+ GDG
Sbjct: 295 IVSHYGIDNYTSIGDAIAAAPNNTKPEDGYFLVYVREGLYEEYVVIPKEKKNILLVGDGI 474
Query: 317 ARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSAL 376
+TI+TG S G T ++TFA + FI +TF NTAGP+ HQAVA RN D+S
Sbjct: 475 NKTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDVTFRNTAGPEKHQAVAVRNNADLSTF 654
Query: 377 VGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTIT 436
C GYQD+LYV S RQ+YR C + GTVDFIFG++A + Q I RKP Q N +T
Sbjct: 655 YRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAAVVFQGCKIYARKPLPNQKNAVT 834
Query: 437 ADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHP 496
A G N NTGI IQ+C+I L + + S+LGRPWK ++TV ++S IG+ I P
Sbjct: 835 AQGRTDPNQNTGISIQNCSIDAAPDLVADLNSTMSFLGRPWKVYSRTVYLQSYIGNVIQP 1014
Query: 497 DGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFT 547
GW W G +T +Y E+ N GPG+N + RV W GY +++ +A FT
Sbjct: 1015AGWLEWNGTVGLDTLFYGEFNNYGPGSNTSNRVTWPGY-SLLNATQAWNFT 1164
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 249 bits (635), Expect = 3e-66
Identities = 133/312 (42%), Positives = 180/312 (57%), Gaps = 6/312 (1%)
Frame = +3
Query: 261 VAKDGSGQFKTIQAALASYPK---GNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPA 317
V++DGSG F TI A+A+ P G ++IYV AGVY+E +++ K ++M GDG
Sbjct: 159 VSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAGVYEENVSIDKKKTYLMMVGDGIN 338
Query: 318 RTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALV 377
+TI+TG +S G T +AT A GF+G MT NTAG HQAVA R+ D+S
Sbjct: 339 KTIITGNRSVVDGWTTFSSATLAVVGQGFVGVNMTIRNTAGAVKHQAVALRSGADLSTFY 518
Query: 378 GCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITA 437
C GYQD+LYV S RQ+Y C + GTVDFIFG++ + Q+ + R P GQFN ITA
Sbjct: 519 SCSFEGYQDTLYVHSLRQFYSECDIYGTVDFIFGNAKVVFQNCKMYPRLPMSGQFNAITA 698
Query: 438 DGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPD 497
G N +TGI I +C I L + +YLGRPWK ++TV M++ + IH
Sbjct: 699 QGRTDPNQDTGISIHNCTIRAADDLAASN-GVATYLGRPWKEYSRTVYMQTVMDSVIHAK 875
Query: 498 GWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAG-- 555
GW W G+ +T YYAEY+N+GPG+ RV W GYH VI+ +A FT +L
Sbjct: 876 GWREWDGDFALSTLYYAEYSNSGPGSGTDNRVTWPGYH-VINATDAANFTVSNFLLGDDW 1052
Query: 556 -PKSAAEWLNGL 566
P++ + N L
Sbjct: 1053LPQTGVSYTNNL 1088
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 238 bits (608), Expect = 4e-63
Identities = 126/272 (46%), Positives = 164/272 (59%)
Frame = +3
Query: 298 EYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTA 357
E I + + N+++ GDG TIVTG + G T ++ATFA GFI + +TFENTA
Sbjct: 9 ENIDIKRTVKNLMIVGDGMGATIVTGNHNAIDGSTTFRSATFAVDGDGFIARDITFENTA 188
Query: 358 GPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLI 417
GP HQAVA R+ D S C GYQD+LYV +NRQ+YR+C + GTVDFIFG + ++
Sbjct: 189 GPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDAVAVL 368
Query: 418 QHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPW 477
Q+ I VRKP Q NT+TA G N NTGI+I +C I L + + R++LGRPW
Sbjct: 369 QNCNIYVRKPMSNQQNTVTAQGRTDPNENTGIIIHNCRITAAGDLKAVQGSFRTFLGRPW 548
Query: 478 KYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGV 537
+ ++TVVM+S + I P GW W G +T YYAE+ANTG GA+ RV W G+ V
Sbjct: 549 QKYSRTVVMKSALDGLISPAGWFPWSGNFALSTLYYAEHANTGAGASTGGRVDWAGFR-V 725
Query: 538 ISRAEANKFTAGIWLQAGPKSAAEWLNGLHVP 569
IS EA KFT G +L G W+ G VP
Sbjct: 726 ISSTEAVKFTVGNFLAGG-----SWIPGSGVP 806
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 234 bits (598), Expect = 6e-62
Identities = 113/251 (45%), Positives = 155/251 (61%), Gaps = 2/251 (0%)
Frame = +1
Query: 305 DAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQA 364
+ N+++ GDG T++TG +F G T +TAT A GFI + + F+NTAGP HQA
Sbjct: 1 EKTNVMLVGDGKDATVITGNLNFIDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQA 180
Query: 365 VAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIV 424
VA R D S + C I +QD+LY SNRQ+YR+ ++GTVDFIFG++A + Q ++
Sbjct: 181 VALRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVA 360
Query: 425 RKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTV 484
RKP Q N +TA G + N NTG IQ CN+ P + L P +I+++LGRPWK ++TV
Sbjct: 361 RKPMDKQNNMVTAQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTFLGRPWKKYSRTV 540
Query: 485 VMESTIGDFIHPDGWTIWQGEQNH--NTCYYAEYANTGPGANVARRVKWKGYHGVISRAE 542
VM+ST+ I P GW W + T YY EY N GPGA ++RV W GYH + + AE
Sbjct: 541 VMQSTLDSHIDPTGWAEWDAQSKDFLQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAAE 720
Query: 543 ANKFTAGIWLQ 553
A+KFT +Q
Sbjct: 721 ASKFTVAQLIQ 753
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 168 bits (426), Expect(2) = 8e-61
Identities = 106/297 (35%), Positives = 155/297 (51%), Gaps = 4/297 (1%)
Frame = +2
Query: 106 KMALNDCKDLMQFALDSLDLS----TKCVHDNNIQAVHDQIADMRNWLSAVISYRQACME 161
K A DC L ++ + L+ + TKC D + WLS ++ + C
Sbjct: 392 KAAWADCLQLYEYTIQRLNKTINPNTKCNQ-----------TDTQTWLSTALTNLETCKN 538
Query: 162 GFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLN 221
GF + + + ++V K+ + L + G + KP S +
Sbjct: 539 GFYELG-----VPDYVLPLMSNNVTKLLSNTLSLNKG----------PYQYKPPSYK--- 664
Query: 222 SEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPK 281
+G+P+W+ DRKLL Q + +N N VVAKDGSG++ T++AA+ + PK
Sbjct: 665 -------EGFPTWVKPGDRKLL---QSSSVASNA--NVVVAKDGSGKYTTVKAAVDAAPK 808
Query: 282 GNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFAN 341
+ GRYVIYVK+GVY+E + V + NI++ GDG +TI+TG KS G T ++AT A
Sbjct: 809 SSSGRYVIYVKSGVYNEQVEVKGN--NIMLVGDGIGKTIITGSKSVGGGTTTFRSATVAA 982
Query: 342 TAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYR 398
GFI + +TF NTAG HQAVAFR+ D+S C G+QD+LYV S RQ+YR
Sbjct: 983 VGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYRCSFEGFQDTLYVHSERQFYR 1153
Score = 84.3 bits (207), Expect(2) = 8e-61
Identities = 41/105 (39%), Positives = 63/105 (59%)
Frame = +3
Query: 400 CLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPE 459
C + GTVDFIFG++A ++Q+ I R P + T+TA G N NTGI+I + +
Sbjct: 1158 CDIYGTVDFIFGNAAAVLQNCNIYARTPPQRTI-TVTAQGRTDPNQNTGIIIHNSKVTGA 1334
Query: 460 AALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQG 504
+ P +++SYLGRPW+ ++TV M++ + I+P GW W G
Sbjct: 1335 SGFNPS--SVKSYLGRPWQKYSRTVFMKTYLDSLINPAGWMEWDG 1463
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 229 bits (585), Expect = 2e-60
Identities = 118/258 (45%), Positives = 162/258 (62%), Gaps = 1/258 (0%)
Frame = +1
Query: 230 GYPSWISSSDRKLL-AKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYV 288
G+P+W+S+ DRKLL A + N+ N +VAKDG+G F TI A+A P + R+V
Sbjct: 157 GFPTWLSTKDRKLLQAAVNETNF------NLLVAKDGTGNFTTIAEAVAVAPNSSATRFV 318
Query: 289 IYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIG 348
I++KAG Y E + V + N++ GDG +T+V ++ G T Q+AT A GFI
Sbjct: 319 IHIKAGAYFENVEVIRKKTNLMFVGDGIGKTVVKASRNVVDGWTTFQSATVAVVGDGFIA 498
Query: 349 KAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDF 408
K +TFEN+AGP HQAVA R+ D SA C V YQD+LYV S RQ+YR+C V GTVDF
Sbjct: 499 KGITFENSAGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDF 678
Query: 409 IFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFT 468
IFG++AT++Q+ + RKP + Q N TA G + N NTGI I +C + A L P +
Sbjct: 679 IFGNAATVLQNCNLYARKPNENQRNLFTAQGREDPNQNTGISILNCKVAAAADLIPVKSQ 858
Query: 469 IRSYLGRPWKYLAKTVVM 486
++YLGRPWK ++TV +
Sbjct: 859 FKNYLGRPWKKYSRTVYL 912
>TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (35%)
Length = 851
Score = 213 bits (542), Expect = 2e-55
Identities = 112/243 (46%), Positives = 142/243 (58%)
Frame = +2
Query: 327 FAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQD 386
F G T TATFA FI + M F NTAGP QAVA D + C I +QD
Sbjct: 2 FVDGTPTFSTATFAVFGRNFIARDMGFRNTAGPQKQQAVALMTSADQAVYYRCQIDAFQD 181
Query: 387 SLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLN 446
SLY SNRQ+YR C + GTVDFIFG+SA ++Q+ I+ R P +GQ N ITA G N+N
Sbjct: 182 SLYAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNIMPRVPMQGQQNPITAQGKTDPNMN 361
Query: 447 TGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQ 506
TGI IQ+CNI P F + ++++YLGRPWK + V M S +G FIHP+GW W G
Sbjct: 362 TGISIQNCNITP----FGDLSSVKTYLGRPWKNYSTPVFM*SPMGIFIHPNGWLPWVGNS 529
Query: 507 NHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGL 566
+T +YAE+ N GPGA+ RV WKG VI+R +A+ FT +L S W+
Sbjct: 530 APDTIFYAEFQNVGPGASTKNRVNWKGLR-VITRKQASMFTVKAFL-----SGERWITAS 691
Query: 567 HVP 569
P
Sbjct: 692 GAP 700
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 210 bits (534), Expect = 2e-54
Identities = 106/250 (42%), Positives = 148/250 (58%), Gaps = 3/250 (1%)
Frame = +3
Query: 320 IVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGC 379
I++G +S G T ++ATFA + GFI + ++F+NTAGP+ HQAVA R+ D+S C
Sbjct: 3 IISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRC 182
Query: 380 HIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADG 439
I GYQDSLY + RQ++R C ++GTVD+IFG + + Q+ + V+K Q NTITA G
Sbjct: 183 GIFGYQDSLYTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHG 362
Query: 440 SDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGW 499
N TG Q CNI ++ L P + +SYLGRPWK ++TV M+S + + I +GW
Sbjct: 363 RKDPNEPTGFSFQFCNITADSDLVPWVSSTQSYLGRPWKSYSRTVFMQSYMSEVIRGEGW 542
Query: 500 TIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAG---P 556
W G T YY EY NTG GA +A RVKW GYH +A+ FT +++ P
Sbjct: 543 LEWNGNFALETLYYGEYMNTGAGAGLANRVKWPGYHPFNDSNQASNFTVAQFIEGNLWLP 722
Query: 557 KSAAEWLNGL 566
+ + GL
Sbjct: 723 STGVTYTAGL 752
>TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (39%)
Length = 808
Score = 207 bits (527), Expect = 1e-53
Identities = 112/245 (45%), Positives = 145/245 (58%), Gaps = 1/245 (0%)
Frame = +2
Query: 320 IVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGC 379
++TG F A VK GFI K + F N AG HQAVAFR+ D S C
Sbjct: 50 VLTGSMLFFAAVK----------GKGFIAKDIGFVNNAGASKHQAVAFRSGSDRSVFFRC 199
Query: 380 HIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADG 439
G+QD+LY SNRQ+YR+C ++GT+DFIFG++A + Q+ I+ R+P QFNTITA G
Sbjct: 200 SFNGFQDTLYAHSNRQFYRDCDITGTIDFIFGNAAAVFQNCKIMPRQPLPNQFNTITAQG 379
Query: 440 SDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGW 499
N NTGI+IQ P T +YLGRPWK + TV+M+S IG F+ P GW
Sbjct: 380 KKDRNQNTGIIIQKSKFTP----LENNLTAPTYLGRPWKDFSTTVIMQSDIGSFLKPVGW 547
Query: 500 TIW-QGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKS 558
W + +T +YAEY NTGPGA+V++RVKW GY ++ EA KFT ++Q GP
Sbjct: 548 MSWVPNVEPVSTIFYAEYQNTGPGADVSQRVKWAGYKPTLTDGEAGKFTVQSFIQ-GP-- 718
Query: 559 AAEWL 563
EWL
Sbjct: 719 --EWL 727
>AW164237
Length = 415
Score = 122 bits (305), Expect(2) = 1e-53
Identities = 56/80 (70%), Positives = 68/80 (85%)
Frame = +2
Query: 391 QSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIV 450
++NRQ+YRNC++SGTVDFIFG+S T+IQHS IIVRKP QFNTITADG+ N++TGIV
Sbjct: 176 RTNRQFYRNCVISGTVDFIFGTSPTVIQHSVIIVRKPLDNQFNTITADGTSMKNMDTGIV 355
Query: 451 IQDCNIIPEAALFPERFTIR 470
IQ CNIIPEA LFP RF ++
Sbjct: 356 IQGCNIIPEAELFPTRFQVK 415
Score = 107 bits (266), Expect(2) = 1e-53
Identities = 50/62 (80%), Positives = 56/62 (89%)
Frame = +1
Query: 333 TMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQS 392
T QTATFANTA GFI KAMTF+NTAG +GHQAVAFRNQGD SALVGCHI+GYQD+LYVQ+
Sbjct: 1 TRQTATFANTAEGFIAKAMTFQNTAGAEGHQAVAFRNQGDRSALVGCHILGYQDTLYVQN 180
Query: 393 NR 394
+
Sbjct: 181 QQ 186
>AW733850
Length = 424
Score = 205 bits (521), Expect = 5e-53
Identities = 93/137 (67%), Positives = 112/137 (80%)
Frame = +1
Query: 368 RNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKP 427
RNQGDMSA+ C + GYQD+LYV +NRQ+YRNC +SGT+DFIFG+SATLIQ+S +IVRKP
Sbjct: 7 RNQGDMSAMFDCAMHGYQDTLYVHANRQFYRNCEISGTIDFIFGASATLIQNSRVIVRKP 186
Query: 428 GKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVME 487
QFNT+TADG+ N+ TGIV+Q+C I+PE ALFP RF +SYLGRPWK A+TVVME
Sbjct: 187 EANQFNTVTADGTKQKNMATGIVLQNCEILPEQALFPSRFQTKSYLGRPWKEFARTVVME 366
Query: 488 STIGDFIHPDGWTIWQG 504
S IGDFI P+GWT W G
Sbjct: 367 SNIGDFIQPEGWTPWDG 417
>TC231526 similar to UP|PME_MEDSA (Q42920) Pectinesterase precursor (Pectin
methylesterase) (PE) (P65) , partial (56%)
Length = 966
Score = 192 bits (487), Expect = 4e-49
Identities = 103/257 (40%), Positives = 147/257 (57%), Gaps = 4/257 (1%)
Frame = +2
Query: 321 VTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCH 380
+TG K++ GV+T TATF A F+ K + FENTAG + HQAVA R D + C+
Sbjct: 8 ITGSKNYVDGVQTYNTATFGVNAANFMAKNIGFENTAGAEKHQAVALRVTADKAVFYNCN 187
Query: 381 IVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGS 440
+ G+QD+LY QS RQ+YR+C V+GT+DF+FG + + Q+ IVR P + Q +TA G
Sbjct: 188 MDGFQDTLYTQSQRQFYRDCTVTGTIDFVFGDAVAVFQNCKFIVRMPLENQQCLVTAGGR 367
Query: 441 DTMNLNTGIVIQDCNIIPE---AALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPD 497
++ + +V Q C E AL P+ +YLGRPW+ AK V+M+S I D P+
Sbjct: 368 SKIDSPSALVFQSCVFTGEPNVLALTPK----IAYLGRPWRLYAKVVIMDSQIDDIFVPE 535
Query: 498 GWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQ-AGP 556
G+ W G +T Y E+ N GPGAN R+ W G+ V++ EA ++ G + Q A
Sbjct: 536 GYMAWMGSAFKDTSTYYEFNNRGPGANTIGRITWPGFK-VLNPIEAVEYYPGKFFQIANS 712
Query: 557 KSAAEWLNGLHVPHYLG 573
W+ G VP+ LG
Sbjct: 713 TERDSWILGSGVPYSLG 763
>TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (39%)
Length = 601
Score = 191 bits (486), Expect = 6e-49
Identities = 94/199 (47%), Positives = 125/199 (62%)
Frame = +2
Query: 351 MTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIF 410
+TF NTAG HQAVA R+ D+S C GYQD+LYV S+RQ+YR C + GTVDFIF
Sbjct: 8 ITFRNTAGATNHQAVALRSGSDLSVFYRCSFEGYQDTLYVYSDRQFYRECDIYGTVDFIF 187
Query: 411 GSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIR 470
G++A + Q+ I R P + NTITA G N NTGI I + + + L +R
Sbjct: 188 GNAAVVFQNCNIYARNP-PNKVNTITAQGRTDPNQNTGISIHNSKVTAASDLMG----VR 352
Query: 471 SYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVK 530
+YLGRPW+ ++TV M++ + I+P+GW W G +T YY EY NTGPG++ A RV
Sbjct: 353 TYLGRPWQQYSRTVFMKTYLDSLINPEGWLEWSGNFALSTLYYGEYMNTGPGSSTANRVN 532
Query: 531 WKGYHGVISRAEANKFTAG 549
W GYH + S +EA+KFT G
Sbjct: 533 WLGYHVITSASEASKFTVG 589
>TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (Fragment) ,
partial (49%)
Length = 1026
Score = 190 bits (483), Expect = 1e-48
Identities = 97/211 (45%), Positives = 124/211 (57%)
Frame = +3
Query: 363 QAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTI 422
QAVA R GD+SA C I+ +QD+LYV +NRQ++ CL++GTVDFIFG+SA + Q I
Sbjct: 6 QAVALRVGGDLSAFFNCDILAFQDTLYVHNNRQFFEKCLIAGTVDFIFGNSAVVFQDCDI 185
Query: 423 IVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAK 482
R P GQ N +TA G N NTGIVIQ C I L + ++YLGRPWK ++
Sbjct: 186 HARLPSSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATNDLESVKKNFKTYLGRPWKEYSR 365
Query: 483 TVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAE 542
TV+M+S+I D I P GW W G +T Y EY NTGPGA + RV WKGY + AE
Sbjct: 366 TVIMQSSISDVIDPIGWHEWSGNFALSTLVYREYQNTGPGAGTSNRVTWKGYKVITDAAE 545
Query: 543 ANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
A +T G ++ + WL P LG
Sbjct: 546 ARDYTPGSFI-----GGSSWLGSTGFPFSLG 623
>CK606276
Length = 751
Score = 177 bits (450), Expect = 9e-45
Identities = 95/211 (45%), Positives = 125/211 (59%), Gaps = 3/211 (1%)
Frame = +1
Query: 368 RNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKP 427
RN DMS C GYQD+LYV S RQ+Y++C + GTVDFIFG++A L+Q + R P
Sbjct: 1 RNGADMSTFYNCSFEGYQDTLYVHSLRQFYKSCDIYGTVDFIFGNAAALLQDCNMYPRLP 180
Query: 428 GKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFT---IRSYLGRPWKYLAKTV 484
+ QFN ITA G N NTGI IQ+C II + L I++YLGRPWK ++TV
Sbjct: 181 MQNQFNAITAQGRTDPNQNTGISIQNCCIIAASDLGDATNNYNGIKTYLGRPWKEYSRTV 360
Query: 485 VMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEAN 544
M+S I I P GW W G+ +T YYAE+AN GPG+N + RV W+GYH +I +A+
Sbjct: 361 YMQSFIDGLIDPKGWNEWSGDFALSTLYYAEFANWGPGSNTSNRVAWEGYH-LIDEKDAD 537
Query: 545 KFTAGIWLQAGPKSAAEWLNGLHVPHYLGFK 575
FT ++Q +WL VP G +
Sbjct: 538 DFTVHKFIQ-----GEKWLPQTGVPFXAGLQ 615
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,094,630
Number of Sequences: 63676
Number of extensions: 291357
Number of successful extensions: 1400
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 1332
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1340
length of query: 576
length of database: 12,639,632
effective HSP length: 102
effective length of query: 474
effective length of database: 6,144,680
effective search space: 2912578320
effective search space used: 2912578320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144563.2


