

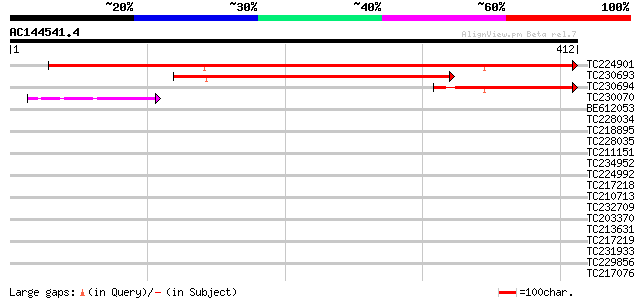
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.4 - phase: 0
(412 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC224901 similar to UP|Q9M4C5 (Q9M4C5) VIP2 protein, partial (23%) 590 e-169
TC230693 homologue to UP|Q6HEM2 (Q6HEM2) Cytochrome c oxidase, s... 296 1e-80
TC230694 127 9e-30
TC230070 weakly similar to GB|AAB68216.1|1244771|SCU43703 Ypl146... 41 9e-04
BE612053 weakly similar to PIR|T51844|T518 RING-H2 finger protei... 41 0.001
TC228034 similar to GB|AAP13413.1|30023760|BT006305 At3g55530 {A... 39 0.003
TC218895 homologue to UP|Q8LJR8 (Q8LJR8) RING-H2 finger protein,... 39 0.003
TC228035 similar to GB|AAP13413.1|30023760|BT006305 At3g55530 {A... 39 0.003
TC211151 UP|Q8LJR8 (Q8LJR8) RING-H2 finger protein, complete 39 0.003
TC234952 weakly similar to UP|Q8L6T7 (Q8L6T7) Ring-H2 zinc finge... 39 0.003
TC224992 39 0.004
TC217218 similar to GB|AAO42794.1|28416527|BT004548 At5g42940/MB... 39 0.006
TC210713 similar to UP|Q8S9I2 (Q8S9I2) At2g38970/T7F6.14, partia... 39 0.006
TC232709 weakly similar to UP|Q9LZJ6 (Q9LZJ6) RING-H2 zinc finge... 38 0.010
TC203370 weakly similar to UP|Q9LX02 (Q9LX02) ESTs AU082316(E336... 38 0.010
TC213631 similar to PIR|F86340|F86340 protein F2D10.34 [imported... 38 0.010
TC217219 similar to GB|AAO42794.1|28416527|BT004548 At5g42940/MB... 38 0.010
TC231933 similar to PIR|F86340|F86340 protein F2D10.34 [imported... 38 0.010
TC229856 similar to UP|Q7Z615 (Q7Z615) Specificity protein 8 sho... 37 0.012
TC217076 similar to UP|Q9SPL2 (Q9SPL2) COP1-interacting protein ... 37 0.012
>TC224901 similar to UP|Q9M4C5 (Q9M4C5) VIP2 protein, partial (23%)
Length = 1796
Score = 590 bits (1520), Expect = e-169
Identities = 280/392 (71%), Positives = 316/392 (80%), Gaps = 8/392 (2%)
Frame = +3
Query: 29 IVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRKIEKGQWLYANGSRSYPEFN 88
+V+DNGDRS++KLQCGHQFHLDCIGSAFN KGAMQCPNCRKIEKGQWLYANG RSYPEF+
Sbjct: 3 VVADNGDRSWSKLQCGHQFHLDCIGSAFNIKGAMQCPNCRKIEKGQWLYANGCRSYPEFS 182
Query: 89 MEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEEREYSPIPYHDAPG--PMFTEH 146
M++WT D+D+YDLSYSEMS GVHWCPFGN A+LPSSFEE E+S YHD G +F EH
Sbjct: 183 MDEWTHDEDLYDLSYSEMSFGVHWCPFGNLARLPSSFEEGEFSSSAYHDVLGQHAIFAEH 362
Query: 147 SAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLH 206
+AVSS SHPCPYIAY GP+HPS+SNSGGTVSE SNFNHWN + DM TSYT PAV LH
Sbjct: 363 TAVSSASHPCPYIAYFGPIHPSSSNSGGTVSEASNFNHWNGSSVPSDMPTSYTFPAVDLH 542
Query: 207 YHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTR-GSEATRSGSYMHPYPVGHSSVARA 265
YHSW+HHS FS+ SS L AADQPSVS +QRP R GS+ RSGS+MHP+ VGHSS AR
Sbjct: 543 YHSWEHHSPPFSTASSRLVAADQPSVSPGSQRPARGGSDVPRSGSFMHPFLVGHSSAARV 722
Query: 266 GNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAP 325
G+S ASSMIPPYPGSNAR RDRV+ALQAYY QQPPNSTT+R P+AS RRSS H+ A
Sbjct: 723 GSSVASSMIPPYPGSNARTRDRVQALQAYYQPQQPPNSTTMRTPIASGTRRSSSHNGPAQ 902
Query: 326 LASLSSPPDQSGSYIYLP----GRNFQEET-RLPSHFHAWERDHLPSSSLNQVGRESSWR 380
LA +++ PDQ G + +P GRNFQEE LP+HFHAWERDHLPS SLN V R+S WR
Sbjct: 903 LAPMATSPDQGGGFFLIPSSSSGRNFQEENHHLPNHFHAWERDHLPSLSLNHVDRDSGWR 1082
Query: 381 AYHQTASGTDPGFRSSSFRLRSDSERTPSQNR 412
AYHQ S +DP RSSSFRLR S+R PSQNR
Sbjct: 1083AYHQATSRSDPATRSSSFRLRHGSDRMPSQNR 1178
>TC230693 homologue to UP|Q6HEM2 (Q6HEM2) Cytochrome c oxidase, subunit III
, partial (6%)
Length = 630
Score = 296 bits (758), Expect = 1e-80
Identities = 146/207 (70%), Positives = 164/207 (78%), Gaps = 3/207 (1%)
Frame = +2
Query: 120 QLPSSFEEREYSPIPYHDAPGP--MFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVS 177
+LPSSFEE ++ I YHD G +F EH+AVSS SH CPYIAY GP+HPSTSNSG TVS
Sbjct: 2 RLPSSFEEGDFYSIAYHDMLGQQAIFAEHTAVSSASHACPYIAYFGPIHPSTSNSGLTVS 181
Query: 178 EVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQ 237
E SNFNHWN P+ GDM TSYT PAV LHYHSW+H+SSHFSS +S LG ++Q SVS +Q
Sbjct: 182 EASNFNHWNGPPVPGDMPTSYTFPAVDLHYHSWEHNSSHFSSANSRLGTSEQTSVSPGSQ 361
Query: 238 RPTR-GSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYH 296
RP R GS+ RSGS+MHP+ VGHSS ARAGNS ASSMIPPYPGSNARARDRV+ALQAYY
Sbjct: 362 RPARGGSDVPRSGSFMHPFLVGHSSAARAGNSVASSMIPPYPGSNARARDRVQALQAYYQ 541
Query: 297 QQQPPNSTTVRPPVASSARRSSGHSRS 323
Q P NS T+R PVASS RRSS HS S
Sbjct: 542 PQLPHNSATIRTPVASSTRRSSSHSGS 622
>TC230694
Length = 709
Score = 127 bits (319), Expect = 9e-30
Identities = 67/108 (62%), Positives = 77/108 (71%), Gaps = 4/108 (3%)
Frame = +1
Query: 309 PVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLP----GRNFQEETRLPSHFHAWERDH 364
P A SAR S LA ++S PD+S ++Y+P G NFQEET LPS FHA ER+H
Sbjct: 1 PRAESARGS------VQLAPVASSPDRSAGFVYIPSGSSGHNFQEETHLPSRFHAREREH 162
Query: 365 LPSSSLNQVGRESSWRAYHQTASGTDPGFRSSSFRLRSDSERTPSQNR 412
LPS LN VGRESSWRAYHQTAS +DP RSSSFRLR +S+R PSQNR
Sbjct: 163 LPSLPLNHVGRESSWRAYHQTASVSDPSIRSSSFRLRHESDRMPSQNR 306
>TC230070 weakly similar to GB|AAB68216.1|1244771|SCU43703 Ypl146cp
{Saccharomyces cerevisiae;} , partial (5%)
Length = 1276
Score = 41.2 bits (95), Expect = 9e-04
Identities = 28/97 (28%), Positives = 51/97 (51%), Gaps = 1/97 (1%)
Frame = +2
Query: 14 HRKSSDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCR-KIEK 72
H K + V C+IC ++++ + + +L C H +H++CI +A+ + CP CR ++
Sbjct: 497 HEKHGELV-CAICKDVLAPRTEVN--QLPCSHLYHINCILPWLSARNS--CPLCRYELPT 661
Query: 73 GQWLYANGSRSYPEFNMEDWTRDDDVYDLSYSEMSLG 109
Y G ++ N+ + DV D SYS++S G
Sbjct: 662 DDKDYEEGKQNIDSRNVIHERQRIDVTDDSYSDVSDG 772
>BE612053 weakly similar to PIR|T51844|T518 RING-H2 finger protein RHA3a
[imported] - Arabidopsis thaliana, partial (25%)
Length = 411
Score = 40.8 bits (94), Expect = 0.001
Identities = 25/63 (39%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Frame = +2
Query: 14 HRKSSDSVP----CSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRK 69
H +S S P C+ICL SD GDR C H+FH+DCI + + CP CR
Sbjct: 185 HSCASPSSPSNNICAICLTEFSD-GDRIRFLPNCNHRFHVDCIDKWLLSHSS--CPTCRN 355
Query: 70 IEK 72
+ K
Sbjct: 356 LLK 364
>TC228034 similar to GB|AAP13413.1|30023760|BT006305 At3g55530 {Arabidopsis
thaliana;} , partial (67%)
Length = 947
Score = 39.3 bits (90), Expect = 0.003
Identities = 21/54 (38%), Positives = 27/54 (49%)
Frame = +3
Query: 15 RKSSDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCR 68
+ S D + CS+CLE V N L C HQFH +CI +G CP C+
Sbjct: 561 KASDDDLTCSVCLEQV--NVGDVLRSLPCLHQFHANCIDPWLRQQGT--CPVCK 710
>TC218895 homologue to UP|Q8LJR8 (Q8LJR8) RING-H2 finger protein, partial
(92%)
Length = 1212
Score = 39.3 bits (90), Expect = 0.003
Identities = 19/47 (40%), Positives = 25/47 (52%)
Frame = +3
Query: 23 CSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRK 69
CSICLE +D + +L CGH+FH C+ G CP CR+
Sbjct: 783 CSICLESFTDGDE--LIRLPCGHKFHSVCLDPWIRCCG--DCPYCRR 911
>TC228035 similar to GB|AAP13413.1|30023760|BT006305 At3g55530 {Arabidopsis
thaliana;} , partial (19%)
Length = 582
Score = 39.3 bits (90), Expect = 0.003
Identities = 21/54 (38%), Positives = 27/54 (49%)
Frame = +3
Query: 15 RKSSDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCR 68
+ S D + CS+CLE V N L C HQFH +CI +G CP C+
Sbjct: 123 KASDDDLTCSVCLEQV--NVGDVLRSLPCLHQFHANCIDPWLRQQGT--CPVCK 272
>TC211151 UP|Q8LJR8 (Q8LJR8) RING-H2 finger protein, complete
Length = 1107
Score = 39.3 bits (90), Expect = 0.003
Identities = 19/47 (40%), Positives = 25/47 (52%)
Frame = +1
Query: 23 CSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRK 69
CSICLE +D + +L CGH+FH C+ G CP CR+
Sbjct: 697 CSICLESFTDGDE--LIRLPCGHKFHSVCLDPWIRCCG--DCPYCRR 825
>TC234952 weakly similar to UP|Q8L6T7 (Q8L6T7) Ring-H2 zinc finger protein,
partial (14%)
Length = 675
Score = 39.3 bits (90), Expect = 0.003
Identities = 18/58 (31%), Positives = 32/58 (55%)
Frame = +2
Query: 11 DTRHRKSSDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCR 68
+ + + +S CS+CL + G+++ + C H++H+DCIG+ K CP CR
Sbjct: 140 EVKDNEEKESDYCSVCLSQIC-KGEKAKSLPVCNHRYHVDCIGAWL--KNHTTCPLCR 304
>TC224992
Length = 615
Score = 38.9 bits (89), Expect = 0.004
Identities = 18/30 (60%), Positives = 20/30 (66%)
Frame = +1
Query: 383 HQTASGTDPGFRSSSFRLRSDSERTPSQNR 412
HQ S +DPG SSFRLR +RT SQNR
Sbjct: 1 HQATSRSDPGXXXSSFRLRHGXDRTXSQNR 90
>TC217218 similar to GB|AAO42794.1|28416527|BT004548 At5g42940/MBD2_14
{Arabidopsis thaliana;} , partial (14%)
Length = 1087
Score = 38.5 bits (88), Expect = 0.006
Identities = 19/51 (37%), Positives = 23/51 (44%)
Frame = +3
Query: 18 SDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCR 68
+D+ PC +C E D D L CGH FH CI K CP C+
Sbjct: 531 TDAEPCCVCQEDYGDGND--IGTLDCGHDFHSSCIKQWLMHKNL--CPICK 671
>TC210713 similar to UP|Q8S9I2 (Q8S9I2) At2g38970/T7F6.14, partial (30%)
Length = 1029
Score = 38.5 bits (88), Expect = 0.006
Identities = 20/58 (34%), Positives = 25/58 (42%), Gaps = 1/58 (1%)
Frame = +1
Query: 12 TRHRKSSDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAK-GAMQCPNCR 68
++ S CSICL + + +C H FH CI S N K G CP CR
Sbjct: 103 SKSSSKSSKQTCSICLTKMKQGSGHAIFTAECSHSFHFHCIAS--NVKHGNQICPVCR 270
>TC232709 weakly similar to UP|Q9LZJ6 (Q9LZJ6) RING-H2 zinc finger protein
ATL5 (At3g62690), partial (19%)
Length = 987
Score = 37.7 bits (86), Expect = 0.010
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Frame = +3
Query: 15 RKSSDSVPCSICL-EIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRKI 70
++ + V CS+CL IV D R C H FH+DC+ FN+ CP CR +
Sbjct: 360 KQGEEVV*CSVCLGTIVEDTISRVLPN--CKHIFHVDCVDKWFNSN--TTCPICRTV 518
>TC203370 weakly similar to UP|Q9LX02 (Q9LX02) ESTs AU082316(E3368), partial
(27%)
Length = 1507
Score = 37.7 bits (86), Expect = 0.010
Identities = 39/129 (30%), Positives = 57/129 (43%), Gaps = 3/129 (2%)
Frame = -3
Query: 206 HYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYM---HPYPVGHSSV 262
H HS + SS S L QPS+S+S+QRP+ AT S S + HP+ S
Sbjct: 662 HPHSAPEKCTSSSSHCSPLFP*SQPSISRSSQRPS----ATSSSSPLLPQHPHSPAEEST 495
Query: 263 ARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSR 322
+R+ SA P P S+ + +Y Q+PP P +S R S
Sbjct: 494 SRSYPSA------PSPASS------YSSSSSYLPPQRPPQ------PTCTSYRTRSSPLP 369
Query: 323 SAPLASLSS 331
+PL++ +S
Sbjct: 368 *SPLSTCTS 342
>TC213631 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(32%)
Length = 589
Score = 37.7 bits (86), Expect = 0.010
Identities = 19/48 (39%), Positives = 27/48 (55%)
Frame = +3
Query: 23 CSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRKI 70
C+ICL + GD QCGH FH+ CI + + + CP+CR+I
Sbjct: 72 CAICLTEFAA-GDEIRVLPQCGHGFHVSCIDAWLRSHSS--CPSCRQI 206
>TC217219 similar to GB|AAO42794.1|28416527|BT004548 At5g42940/MBD2_14
{Arabidopsis thaliana;} , partial (8%)
Length = 454
Score = 37.7 bits (86), Expect = 0.010
Identities = 19/50 (38%), Positives = 22/50 (44%)
Frame = +3
Query: 19 DSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCR 68
D+ PC +C E D D L CGH FH CI K CP C+
Sbjct: 39 DAEPCCVCQEDYGDEDD--IGTLDCGHDFHSSCIKQWLMHKNL--CPICK 176
>TC231933 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(65%)
Length = 619
Score = 37.7 bits (86), Expect = 0.010
Identities = 19/48 (39%), Positives = 27/48 (55%)
Frame = +2
Query: 23 CSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRKI 70
C+ICL + GD QCGH FH+ CI + + + CP+CR+I
Sbjct: 311 CAICLTEFAA-GDEIRVLPQCGHGFHVSCIDAWLRSHSS--CPSCRQI 445
>TC229856 similar to UP|Q7Z615 (Q7Z615) Specificity protein 8 short isoform,
partial (5%)
Length = 821
Score = 37.4 bits (85), Expect = 0.012
Identities = 47/171 (27%), Positives = 67/171 (38%), Gaps = 2/171 (1%)
Frame = +2
Query: 206 HYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPY--PVGHSSVA 263
HYH HH SH ++H+ P+ + PT G AT +H + P SV
Sbjct: 176 HYHHHHHHQSHHH--NTHVFPETSPAPT-----PTTGEGAT-----LHNFGAPAPARSVP 319
Query: 264 RAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRS 323
G S+ + PP N R A+ PP +T PVAS H
Sbjct: 320 APGRSSYAQ--PP----NCRF--------AHVTPAAPP-TTAPHYPVASPWVGPPAHGFD 454
Query: 324 APLASLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVG 374
+ + +LS P+ + ++ P +N L SHFH PS S + G
Sbjct: 455 SSVPALSPLPNVAFAHAEPPPKNEAAAEHLNSHFHG------PSPSSSSAG 589
>TC217076 similar to UP|Q9SPL2 (Q9SPL2) COP1-interacting protein CIP8
(At5g64920), partial (13%)
Length = 575
Score = 37.4 bits (85), Expect = 0.012
Identities = 17/51 (33%), Positives = 32/51 (62%)
Frame = +3
Query: 18 SDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCR 68
S++V C+IC +++ + + +L CGH++H DCI +++ + CP CR
Sbjct: 30 SEAVACAICKDLLGV--EDAAKRLPCGHRYHGDCIVPWLSSRNS--CPVCR 170
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.128 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,710,103
Number of Sequences: 63676
Number of extensions: 394663
Number of successful extensions: 2671
Number of sequences better than 10.0: 214
Number of HSP's better than 10.0 without gapping: 2427
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2624
length of query: 412
length of database: 12,639,632
effective HSP length: 100
effective length of query: 312
effective length of database: 6,272,032
effective search space: 1956873984
effective search space used: 1956873984
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144541.4


