

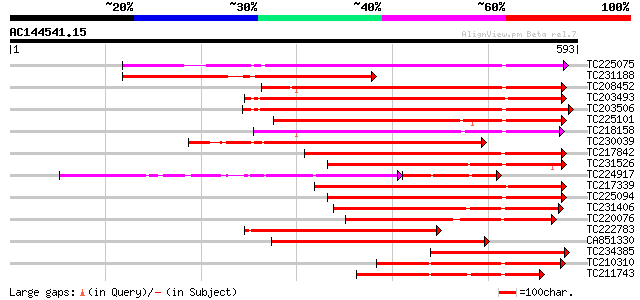
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.15 - phase: 0
(593 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 317 9e-87
TC231188 weakly similar to UP|Q9FJ21 (Q9FJ21) Pectin methylester... 308 3e-84
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 299 3e-81
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 295 4e-80
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 289 3e-78
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 281 6e-76
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 276 2e-74
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 258 7e-69
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 257 9e-69
TC231526 similar to UP|PME_MEDSA (Q42920) Pectinesterase precurs... 257 9e-69
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 178 5e-68
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 246 2e-65
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 242 3e-64
TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (... 222 4e-58
TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 pr... 219 2e-57
TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylester... 217 1e-56
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 208 6e-54
TC234385 similar to UP|Q9FJ21 (Q9FJ21) Pectin methylesterase (AT... 202 3e-52
TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 199 2e-51
TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 190 1e-48
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 317 bits (812), Expect = 9e-87
Identities = 177/469 (37%), Positives = 258/469 (54%), Gaps = 3/469 (0%)
Frame = +1
Query: 119 RTKQALDTCKQVMQLSIDEFQRSLERFSN--FDLNSLDRVLTSLKVWLSGAITYQETCLD 176
R A+ C ++ LS D +L N NS + + L+ WLS A+ + ETC++
Sbjct: 238 RLSTAIADCLDLLDLSSDVLSWALSASQNPKGKHNSTGNLSSDLRTWLSAALAHPETCME 417
Query: 177 AFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDV 236
FE T SI+ L +LL + + ++
Sbjct: 418 GFEGTN----------------------SIVKGLVSAGIGQVVSLVEQLLAQVLPAQDQF 531
Query: 237 LGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNL 296
+ P W+ + RKLL + + V VA DGSGN+ I +A+ P ++
Sbjct: 532 DAASSKGQFPSWIKPKE--RKLLQAIA---VTPDVTVALDGSGNYAKIMDAVLAAPDYSM 696
Query: 297 KPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGD 356
K FVI +K+GVY E VE+ K +++ +G G T I+GN++ +DG TF++A+ A++G
Sbjct: 697 KRFVILVKKGVYVENVEIKKKKWNIMILGQGMDATVISGNRSVVDGWTTFRSATFAVSGR 876
Query: 357 FFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISG 416
F+ I F+N+AGPEKHQAVALR SD S+F++C + GYQD+LY HTMRQF+RDC ISG
Sbjct: 877 GFIARDISFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDCTISG 1056
Query: 417 TIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYP 476
T+D++FGD+ AV QNC V+K L NQ+ +TA GRK+ N+PTG Q +I AD P
Sbjct: 1057TVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHGRKDPNEPTGFSFQFCNITADSDLIP 1236
Query: 477 VRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGP 536
+ YL RPWK +SRT+F+ +Y+ ++I EG++ W G DT YY EY N G
Sbjct: 1237SVGTAQTYLGRPWKSYSRTVFMQSYMSEVIGAEGWLEWN---GNFALDTLYYAEYMNTGA 1407
Query: 537 GSDVKQRVKWQGVKTIT-SEGAASFVPIRFFHGDDWIRVTRVPYSPGAT 584
G+ V RVKW G + S A++F +F G+ W+ T V ++ G T
Sbjct: 1408GAGVANRVKWPGYHALNDSSQASNFTVSQFIEGNLWLPSTGVTFTAGLT 1554
>TC231188 weakly similar to UP|Q9FJ21 (Q9FJ21) Pectin methylesterase
(AT5g49180/K21P3_5), partial (23%)
Length = 758
Score = 308 bits (790), Expect = 3e-84
Identities = 162/268 (60%), Positives = 200/268 (74%), Gaps = 3/268 (1%)
Frame = +1
Query: 119 RTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAF 178
R K ALDTCKQ+M LSI E RSL+ + F+L ++D++L +LKVWLSGA+TYQ+TCLD F
Sbjct: 7 RAKMALDTCKQLMDLSIGELTRSLDGINEFNLINVDKILMNLKVWLSGAVTYQDTCLDGF 186
Query: 179 ENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEE--MKQPAGRRLLKESVDGEEDV 236
ENTT+DAGKKMK++L MHMSSN L+I+ L+ T + + + GRRLL++S
Sbjct: 187 ENTTSDAGKKMKDLLTIGMHMSSNALAIVTDLADTVNDWNITKSFGRRLLQDS------- 345
Query: 237 LGHGGDFELPEWVDDRAGVRKLLNKMTGR-KLQAHVVVAKDGSGNFTTITEALKHVPKKN 295
ELP WVD +LLN+ K + +V VA DGSG+F +I EALK VP+KN
Sbjct: 346 -------ELPSWVDQH----RLLNENASPFKRKPNVTVAIDGSGDFKSINEALKQVPEKN 492
Query: 296 LKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITG 355
KPFVIYIKEGVY+EYVEVTK MTHVVFIG+GG+KTRI+GNKNFIDG T++TA+VAI G
Sbjct: 493 RKPFVIYIKEGVYQEYVEVTKKMTHVVFIGEGGKKTRISGNKNFIDGTNTYRTATVAIQG 672
Query: 356 DFFVGIGIGFENSAGPEKHQAVALRVQS 383
D FV I +GFENSAGP KHQAVALRVQ+
Sbjct: 673 DHFVAINMGFENSAGPHKHQAVALRVQA 756
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 299 bits (765), Expect = 3e-81
Identities = 145/323 (44%), Positives = 208/323 (63%), Gaps = 4/323 (1%)
Frame = +1
Query: 264 GRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKP----FVIYIKEGVYKEYVEVTKTMT 319
G L V+V+ G N+T+I +A+ P N KP F++Y++EG+Y+EYV + K
Sbjct: 271 GILLYDFVIVSHYGIDNYTSIGDAIAAAPN-NTKPEDGYFLVYVREGLYEEYVVIPKEKK 447
Query: 320 HVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVAL 379
+++ +GDG KT ITGN + IDG TF +++ A++G+ F+ + + F N+AGPEKHQAVA+
Sbjct: 448 NILLVGDGINKTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDVTFRNTAGPEKHQAVAV 627
Query: 380 RVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKP 439
R +D S FY+C +GYQDTLY H++RQFYR+C I GT+DF+FG++ V Q C RKP
Sbjct: 628 RNNADLSTFYRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAAVVFQGCKIYARKP 807
Query: 440 LENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLD 499
L NQ+ VTAQGR + NQ TG+ IQ SI A P ++L RPWK +SRT++L
Sbjct: 808 LPNQKNAVTAQGRTDPNQNTGISIQNCSIDAAPDLVADLNSTMSFLGRPWKVYSRTVYLQ 987
Query: 500 TYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAAS 559
+YIG++I P G++ W G G DT +YGE+NN GPGS+ RV W G + + A +
Sbjct: 988 SYIGNVIQPAGWLEWN---GTVGLDTLFYGEFNNYGPGSNTSNRVTWPGYSLLNATQAWN 1158
Query: 560 FVPIRFFHGDDWIRVTRVPYSPG 582
F + F G+ W+ T +PY+ G
Sbjct: 1159FTVLNFTLGNTWLPDTDIPYTEG 1227
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 295 bits (755), Expect = 4e-80
Identities = 153/339 (45%), Positives = 216/339 (63%), Gaps = 2/339 (0%)
Frame = +1
Query: 246 PEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKE 305
P W++ R R+LL+ + ++QA +VV+KDG+G TI EA+K VP+ + + +IYI+
Sbjct: 142 PTWLNGRD--RRLLS-LPLSQIQADIVVSKDGNGTVKTIAEAIKKVPEYSSRRIIIYIRA 312
Query: 306 GVYKE-YVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIG 364
G Y+E +++ + T+V+FIGDG KT ITG +N+ + TF TAS A +G F+ +
Sbjct: 313 GRYEEDNLKLGRKKTNVMFIGDGKGKTVITGGRNYYQNLTTFHTASFAASGSGFIAKDMT 492
Query: 365 FENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGD 424
FEN AGP +HQAVALRV +D ++ Y+C + GYQDT+Y H+ RQFYR+C I GT+DF+FG+
Sbjct: 493 FENYAGPGRHQAVALRVGADHAVVYRCNIIGYQDTMYVHSNRQFYRECDIYGTVDFIFGN 672
Query: 425 SIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAY 484
+ V QNCT RKP+ Q+ +TAQ RK+ NQ TG+ I I+A P + Y
Sbjct: 673 AAVVFQNCTLWARKPMAQQKNTITAQNRKDPNQNTGISIHNCRIMATPDLEASKGSYPTY 852
Query: 485 LARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRV 544
L RPWK ++RT+F+ +YIGD + P G++ W T + DTCYYGEY N GPGS + QRV
Sbjct: 853 LGRPWKLYARTVFMLSYIGDHVHPRGWLEWNTSS--FALDTCYYGEYMNYGPGSALGQRV 1026
Query: 545 KWQGVKTITSEGAAS-FVPIRFFHGDDWIRVTRVPYSPG 582
W G + I S AS F +F G W+ T V + G
Sbjct: 1027NWAGYRAINSTVEASRFTVGQFISGSSWLPSTGVAFIAG 1143
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 289 bits (739), Expect = 3e-78
Identities = 153/350 (43%), Positives = 218/350 (61%), Gaps = 4/350 (1%)
Frame = +2
Query: 244 ELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYI 303
+ P W++ R R+LL+ + +QA + V+K G+G TI EA+K P+ + + F+IY+
Sbjct: 8 KFPGWLNKRD--RRLLS-LPLSAIQADITVSKTGNGTVKTIAEAIKKAPEHSTRRFIIYV 178
Query: 304 KEGVYKEY-VEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
+ G Y+E ++V + T+V+FIGDG KT ITG KN IDG+ TF +AS A +G F+
Sbjct: 179 RAGRYEENNLKVGRKKTNVMFIGDGKGKTVITGKKNVIDGMTTFHSASFAASGAGFIARD 358
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
I FEN AGP KHQAVALRV +D ++ Y+C + GYQD+ Y H+ RQF+R+C I GT+DF+F
Sbjct: 359 ITFENYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFIF 538
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
G++ V Q C RKP+ Q+ +TAQ RK+ NQ TG+ + I+ P PV+
Sbjct: 539 GNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISLHNCRILPAPDLAPVKGSFP 718
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
YL RPWK +SRT++L +Y+GD I P G++ W G +T YYGEY N GPG+ V Q
Sbjct: 719 TYLGRPWKQYSRTVYLVSYMGDHIHPRGWLEWN---GDFALNTLYYGEYMNYGPGAAVGQ 889
Query: 543 RVKWQGVKTITSEGAAS-FVPIRFFHGDDWIRVTRVPYSPGATK--TPPS 589
RV+W G + I S A+ F +F G W+ T V ++ G + TP S
Sbjct: 890 RVQWPGYRVIKSTMEANRFTVAQFISGSAWLPSTGVAFAAGLST*ITPQS 1039
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 281 bits (719), Expect = 6e-76
Identities = 145/312 (46%), Positives = 202/312 (64%), Gaps = 6/312 (1%)
Frame = +3
Query: 277 GSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGN 336
G N+T + +A+ P +++ +VI+IK GVY E VE+ K +++ +GDG T I+GN
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGDGMDATIISGN 386
Query: 337 KNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGY 396
++FIDG TF++A+ A++G F+ I F+N+AGPEKHQAVALR SD S+F++C + GY
Sbjct: 387 RSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGY 566
Query: 397 QDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKN 456
QD+LY HTMRQFYR+C ISGT+DF+FGD+ A+ QNC +K L NQ+ +TA GRK +
Sbjct: 567 QDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAHGRKNPD 746
Query: 457 QPTGLIIQGGSIVADPKYYPVRLKNK-----AYLARPWKDFSRTIFLDTYIGDMITPEGY 511
+PTG IQ +I AD Y V N YL RPWK +SRTIF+ +YI D++ PEG+
Sbjct: 747 EPTGFSIQFCNISAD--YDLVNSVNSCNSTHTYLGRPWKPYSRTIFMQSYISDVLRPEGW 920
Query: 512 MPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTIT-SEGAASFVPIRFFHGDD 570
+ W G DT YY EY N GPG+ V RVKWQG + S A++F +F G+
Sbjct: 921 LEWN---GDFALDTLYYAEYMNYGPGAGVANRVKWQGYHVMNDSSQASNFTVSQFIEGNL 1091
Query: 571 WIRVTRVPYSPG 582
+ T V ++ G
Sbjct: 1092CLPSTGVTFTAG 1127
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 276 bits (705), Expect = 2e-74
Identities = 144/332 (43%), Positives = 199/332 (59%), Gaps = 7/332 (2%)
Frame = +3
Query: 256 RKLLNKMTGRKLQAH--VVVAKDGSGNFTTITEALKHVPKKNLKP---FVIYIKEGVYKE 310
RKLL G ++ V V++DGSGNFTTI +A+ P K++ F+IY+ GVY+E
Sbjct: 102 RKLLQAKVGDEVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAGVYEE 281
Query: 311 YVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAG 370
V + K T+++ +GDG KT ITGN++ +DG TF +A++A+ G FVG+ + N+AG
Sbjct: 282 NVSIDKKKTYLMMVGDGINKTIITGNRSVVDGWTTFSSATLAVVGQGFVGVNMTIRNTAG 461
Query: 371 PEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQ 430
KHQAVALR +D S FY C +GYQDTLY H++RQFY +C I GT+DF+FG++ V Q
Sbjct: 462 AVKHQAVALRSGADLSTFYSCSFEGYQDTLYVHSLRQFYSECDIYGTVDFIFGNAKVVFQ 641
Query: 431 NCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKN--KAYLARP 488
NC R P+ Q +TAQGR + NQ TG+ I +I A + N YL RP
Sbjct: 642 NCKMYPRLPMSGQFNAITAQGRTDPNQDTGISIHNCTIRAAD---DLAASNGVATYLGRP 812
Query: 489 WKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQG 548
WK++SRT+++ T + +I +G+ W G T YY EY+N GPGS RV W G
Sbjct: 813 WKEYSRTVYMQTVMDSVIHAKGWREWD---GDFALSTLYYAEYSNSGPGSGTDNRVTWPG 983
Query: 549 VKTITSEGAASFVPIRFFHGDDWIRVTRVPYS 580
I + AA+F F GDDW+ T V Y+
Sbjct: 984 YHVINATDAANFTVSNFLLGDDWLPQTGVSYT 1079
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 258 bits (658), Expect = 7e-69
Identities = 142/312 (45%), Positives = 194/312 (61%), Gaps = 1/312 (0%)
Frame = +1
Query: 188 KMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFE-LP 246
+ +E L H SN L+++ +L PAG +K+ E G+G + P
Sbjct: 22 RFEEGLLEISHHVSNSLAMLKKL---------PAG---VKKLASKNEVFPGYGKIKDGFP 165
Query: 247 EWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEG 306
W+ + RKLL + +++VAKDG+GNFTTI EA+ P + FVI+IK G
Sbjct: 166 TWLSTKD--RKLLQAAVN-ETNFNLLVAKDGTGNFTTIAEAVAVAPNSSATRFVIHIKAG 336
Query: 307 VYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFE 366
Y E VEV + T+++F+GDG KT + ++N +DG TF++A+VA+ GD F+ GI FE
Sbjct: 337 AYFENVEVIRKKTNLMFVGDGIGKTVVKASRNVVDGWTTFQSATVAVVGDGFIAKGITFE 516
Query: 367 NSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSI 426
NSAGP KHQAVALR SD S FYKC YQDTLY H++RQFYRDC + GT+DF+FG++
Sbjct: 517 NSAGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDFIFGNAA 696
Query: 427 AVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLA 486
VLQNC RKP ENQ+ + TAQGR++ NQ TG+ I + A PV+ + K YL
Sbjct: 697 TVLQNCNLYARKPNENQRNLFTAQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLG 876
Query: 487 RPWKDFSRTIFL 498
RPWK +SRT++L
Sbjct: 877 RPWKKYSRTVYL 912
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 257 bits (657), Expect = 9e-69
Identities = 125/274 (45%), Positives = 173/274 (62%)
Frame = +3
Query: 309 KEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENS 368
KE +++ +T+ +++ +GDG T +TGN N IDG TF++A+ A+ GD F+ I FEN+
Sbjct: 6 KENIDIKRTVKNLMIVGDGMGATIVTGNHNAIDGSTTFRSATFAVDGDGFIARDITFENT 185
Query: 369 AGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAV 428
AGP+KHQAVALR +D S+FY+C GYQDTLY + RQFYRDC I GT+DF+FGD++AV
Sbjct: 186 AGPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDAVAV 365
Query: 429 LQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARP 488
LQNC VRKP+ NQQ VTAQGR + N+ TG+II I A V+ + +L RP
Sbjct: 366 LQNCNIYVRKPMSNQQNTVTAQGRTDPNENTGIIIHNCRITAAGDLKAVQGSFRTFLGRP 545
Query: 489 WKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQG 548
W+ +SRT+ + + + +I+P G+ PW +G T YY E+ N G G+ RV W G
Sbjct: 546 WQKYSRTVVMKSALDGLISPAGWFPW---SGNFALSTLYYAEHANTGAGASTGGRVDWAG 716
Query: 549 VKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
+ I+S A F F G WI + VP+ G
Sbjct: 717 FRVISSTEAVKFTVGNFLAGGSWIPGSGVPFDEG 818
>TC231526 similar to UP|PME_MEDSA (Q42920) Pectinesterase precursor (Pectin
methylesterase) (PE) (P65) , partial (56%)
Length = 966
Score = 257 bits (657), Expect = 9e-69
Identities = 124/256 (48%), Positives = 166/256 (64%), Gaps = 6/256 (2%)
Frame = +2
Query: 333 ITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCR 392
ITG+KN++DGV T+ TA+ + F+ IGFEN+AG EKHQAVALRV +D+++FY C
Sbjct: 8 ITGSKNYVDGVQTYNTATFGVNAANFMAKNIGFENTAGAEKHQAVALRVTADKAVFYNCN 187
Query: 393 MDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGR 452
MDG+QDTLY + RQFYRDC ++GTIDFVFGD++AV QNC F+VR PLENQQC+VTA GR
Sbjct: 188 MDGFQDTLYTQSQRQFYRDCTVTGTIDFVFGDAVAVFQNCKFIVRMPLENQQCLVTAGGR 367
Query: 453 KEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYM 512
+ + P+ L+ Q +P + K AYL RPW+ +++ + +D+ I D+ PEGYM
Sbjct: 368 SKIDSPSALVFQSCVFTGEPNVLALTPK-IAYLGRPWRLYAKVVIMDSQIDDIFVPEGYM 544
Query: 513 PWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAASFVPIRFF------ 566
W G DT Y E+NNRGPG++ R+ W G K + A + P +FF
Sbjct: 545 AWM---GSAFKDTSTYYEFNNRGPGANTIGRITWPGFKVLNPIEAVEYYPGKFFQIANST 715
Query: 567 HGDDWIRVTRVPYSPG 582
D WI + VPYS G
Sbjct: 716 ERDSWILGSGVPYSLG 763
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 178 bits (452), Expect(2) = 5e-68
Identities = 125/360 (34%), Positives = 177/360 (48%), Gaps = 2/360 (0%)
Frame = +2
Query: 53 SQSVKAVKTLCAPTDYKKECEDSLIAHAGN--ITEPKELIKIAFNITIAKISEGLKKTHL 110
S S K +++ C T Y + CE L HA N I + +K++ + + + TH
Sbjct: 182 SYSFKDIQSWCNQTPYPQPCEYYLTNHAFNKPIKSKSDFLKVSLQLALERAQRSELNTHA 361
Query: 111 LQEAEKDERTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITY 170
L ++ K A C Q+ + +I +++ N N D + WLS A+T
Sbjct: 362 LGPKCRNVHEKAAWADCLQLYEYTIQRLNKTIN--PNTKCNQTDT-----QTWLSTALTN 520
Query: 171 QETCLDAFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESV 230
ETC + F VL + + LS L+K + K P+ KE
Sbjct: 521 LETCKNGFYELGVP-----DYVLPLMSNNVTKLLSNTLSLNKGPYQYKPPS----YKEG- 670
Query: 231 DGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKH 290
P WV + G RKLL + A+VVVAKDGSG +TT+ A+
Sbjct: 671 --------------FPTWV--KPGDRKLLQSSSVAS-NANVVVAKDGSGKYTTVKAAVDA 799
Query: 291 VPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTAS 350
PK + +VIY+K GVY E VEV +++ +GDG KT ITG+K+ G TF++A+
Sbjct: 800 APKSSSGRYVIYVKSGVYNEQVEVKGN--NIMLVGDGIGKTIITGSKSVGGGTTTFRSAT 973
Query: 351 VAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYR 410
VA GD F+ I F N+AG HQAVA R SD S+FY+C +G+QDTLY H+ RQFYR
Sbjct: 974 VAAVGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYRCSFEGFQDTLYVHSERQFYR 1153
Score = 98.6 bits (244), Expect(2) = 5e-68
Identities = 46/104 (44%), Positives = 66/104 (63%)
Frame = +3
Query: 411 DCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVA 470
+C I GT+DF+FG++ AVLQNC R P + + VTAQGR + NQ TG+II +
Sbjct: 1155 ECDIYGTVDFIFGNAAAVLQNCNIYARTPPQ-RTITVTAQGRTDPNQNTGIIIHNSKVTG 1331
Query: 471 DPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPW 514
+ P +K+ YL RPW+ +SRT+F+ TY+ +I P G+M W
Sbjct: 1332 ASGFNPSSVKS--YLGRPWQKYSRTVFMKTYLDSLINPAGWMEW 1457
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 246 bits (628), Expect = 2e-65
Identities = 123/265 (46%), Positives = 167/265 (62%), Gaps = 1/265 (0%)
Frame = +1
Query: 319 THVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVA 378
T+V+ +GDG T ITGN NFIDG TFKTA+VA GD F+ I F+N+AGP+KHQAVA
Sbjct: 7 TNVMLVGDGKDATVITGNLNFIDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQAVA 186
Query: 379 LRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRK 438
LRV +D+S+ +CR+D +QDTLYAH+ RQFYRD I+GT+DF+FG++ V Q C V RK
Sbjct: 187 LRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVARK 366
Query: 439 PLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFL 498
P++ Q +VTAQGR++ NQ TG IQ ++ PV K +L RPWK +SRT+ +
Sbjct: 367 PMDKQNNMVTAQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTFLGRPWKKYSRTVVM 546
Query: 499 DTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTI-TSEGA 557
+ + I P G+ W + T YYGEY N GPG+ +RV W G I T+ A
Sbjct: 547 QSTLDSHIDPTGWAEWDAQSK-DFLQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAAEA 723
Query: 558 ASFVPIRFFHGDDWIRVTRVPYSPG 582
+ F + G+ W++ T V + G
Sbjct: 724 SKFTVAQLIQGNVWLKNTGVNFIEG 798
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 242 bits (618), Expect = 3e-64
Identities = 115/251 (45%), Positives = 166/251 (65%), Gaps = 1/251 (0%)
Frame = +3
Query: 333 ITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCR 392
I+GN++ +DG TF++A+ A++G F+ I F+N+AGPEKHQAVALR +D S+F++C
Sbjct: 6 ISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRCG 185
Query: 393 MDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGR 452
+ GYQD+LY HTMRQF+R+C I+GT+D++FGD+ AV QNC V+K L NQ+ +TA GR
Sbjct: 186 IFGYQDSLYTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHGR 365
Query: 453 KEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYM 512
K+ N+PTG Q +I AD P ++YL RPWK +SRT+F+ +Y+ ++I EG++
Sbjct: 366 KDPNEPTGFSFQFCNITADSDLVPWVSSTQSYLGRPWKSYSRTVFMQSYMSEVIRGEGWL 545
Query: 513 PWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTIT-SEGAASFVPIRFFHGDDW 571
W G +T YYGEY N G G+ + RVKW G S A++F +F G+ W
Sbjct: 546 EWN---GNFALETLYYGEYMNTGAGAGLANRVKWPGYHPFNDSNQASNFTVAQFIEGNLW 716
Query: 572 IRVTRVPYSPG 582
+ T V Y+ G
Sbjct: 717 LPSTGVTYTAG 749
>TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (35%)
Length = 851
Score = 222 bits (565), Expect = 4e-58
Identities = 104/241 (43%), Positives = 154/241 (63%)
Frame = +2
Query: 339 FIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQD 398
F+DG TF TA+ A+ G F+ +GF N+AGP+K QAVAL +D++++Y+C++D +QD
Sbjct: 2 FVDGTPTFSTATFAVFGRNFIARDMGFRNTAGPQKQQAVALMTSADQAVYYRCQIDAFQD 181
Query: 399 TLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQP 458
+LYAH+ RQFYR+C I GT+DF+FG+S VLQNC + R P++ QQ +TAQG+ + N
Sbjct: 182 SLYAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNIMPRVPMQGQQNPITAQGKTDPNMN 361
Query: 459 TGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPA 518
TG+ IQ +I V K YL RPWK++S +F+ + +G I P G++PW
Sbjct: 362 TGISIQNCNITPFGDLSSV----KTYLGRPWKNYSTPVFM*SPMGIFIHPNGWLPW---V 520
Query: 519 GITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVP 578
G + DT +Y E+ N GPG+ K RV W+G++ IT + A+ F F G+ WI + P
Sbjct: 521 GNSAPDTIFYAEFQNVGPGASTKNRVNWKGLRVITRKQASMFTVKAFLSGERWITASGAP 700
Query: 579 Y 579
+
Sbjct: 701 F 703
>TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (39%)
Length = 808
Score = 219 bits (559), Expect = 2e-57
Identities = 110/224 (49%), Positives = 143/224 (63%), Gaps = 3/224 (1%)
Frame = +2
Query: 352 AITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRD 411
A+ G F+ IGF N+AG KHQAVA R SDRS+F++C +G+QDTLYAH+ RQFYRD
Sbjct: 80 AVKGKGFIAKDIGFVNNAGASKHQAVAFRSGSDRSVFFRCSFNGFQDTLYAHSNRQFYRD 259
Query: 412 CIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVAD 471
C I+GTIDF+FG++ AV QNC + R+PL NQ +TAQG+K++NQ TG+IIQ
Sbjct: 260 CDITGTIDFIFGNAAAVFQNCKIMPRQPLPNQFNTITAQGKKDRNQNTGIIIQ------K 421
Query: 472 PKYYPV--RLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYG 529
K+ P+ L YL RPWKDFS T+ + + IG + P G+M W + T +Y
Sbjct: 422 SKFTPLENNLTAPTYLGRPWKDFSTTVIMQSDIGSFLKPVGWMSW--VPNVEPVSTIFYA 595
Query: 530 EYNNRGPGSDVKQRVKWQGVK-TITSEGAASFVPIRFFHGDDWI 572
EY N GPG+DV QRVKW G K T+T A F F G +W+
Sbjct: 596 EYQNTGPGADVSQRVKWAGYKPTLTDGEAGKFTVQSFIQGPEWL 727
>TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylesterase
precursor , partial (29%)
Length = 630
Score = 217 bits (552), Expect = 1e-56
Identities = 108/206 (52%), Positives = 138/206 (66%)
Frame = +3
Query: 246 PEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKE 305
P WV A RKLL ++ + H VAKDGSG FTT+ +A+ PKK+ ++IY+K
Sbjct: 18 PTWVS--AADRKLLAQLNDGAVLPHATVAKDGSGQFTTVLDAINSYPKKHQGRYIIYVKA 191
Query: 306 GVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGF 365
G+Y EY+ V K +++ GDG KT ITG KNF +G T +TA+ + + F+ I F
Sbjct: 192 GIYDEYITVDKKKPNLLIYGDGPTKTIITGRKNFHEGTKTMRTATFSTVAEDFMAKSIAF 371
Query: 366 ENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDS 425
EN+AG E HQAVALRVQ DRS+F+ C M GYQDTLYAH RQFYR+C ISGTIDF+FG S
Sbjct: 372 ENTAGAEGHQAVALRVQGDRSVFFDCAMRGYQDTLYAHAHRQFYRNCEISGTIDFIFGYS 551
Query: 426 IAVLQNCTFVVRKPLENQQCIVTAQG 451
++QN +VRKP+ NQQ IV A G
Sbjct: 552 TTLIQNSKILVRKPMPNQQNIVVADG 629
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 208 bits (529), Expect = 6e-54
Identities = 106/229 (46%), Positives = 144/229 (62%), Gaps = 1/229 (0%)
Frame = +2
Query: 275 KDGSGNFTTITEALKHVPKKNLKP-FVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRI 333
K GSGNF T+ +AL K+ K FVI++K+GVY+E +EV +++ +GDG R T I
Sbjct: 8 KXGSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYRENIEVALHNDNIMLVGDGLRNTII 187
Query: 334 TGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRM 393
T ++ DG T+ +A+ I G F+ I F+NSAG K QAVALR SD S+FY+C +
Sbjct: 188 TSARSVQDGYTTYSSATAGIDGLHFIARDITFQNSAGVHKGQAVALRSASDLSVFYRCGI 367
Query: 394 DGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRK 453
GYQDTL AH RQFYR C I GT+DF+FG++ V QNC R+PLE Q ++TAQGR
Sbjct: 368 MGYQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVFQNCYIFARRPLEGQANMITAQGRG 547
Query: 454 EKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYI 502
+ Q TG+ I I A P PV K +L RPW+ +SR + + T++
Sbjct: 548 DPFQNTGISIHNSQIRAAPDLKPVVDKYNTFLGRPWQQYSRVVVMKTFM 694
>TC234385 similar to UP|Q9FJ21 (Q9FJ21) Pectin methylesterase
(AT5g49180/K21P3_5), partial (6%)
Length = 677
Score = 202 bits (515), Expect = 3e-52
Identities = 90/145 (62%), Positives = 112/145 (77%)
Frame = +1
Query: 441 ENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDT 500
ENQQCIVTAQGRKE QP+G++IQGGSIV+DP++Y VR +NKAYLARPWK++SRTI +DT
Sbjct: 16 ENQQCIVTAQGRKEIQQPSGIVIQGGSIVSDPEFYSVRFENKAYLARPWKNYSRTIIMDT 195
Query: 501 YIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAASF 560
YI D+I +GY+PWQ G +G DTC+Y EY+N GPGSD +RVKW G+ + S+ A F
Sbjct: 196 YIDDLIDADGYLPWQGLEGPSGMDTCFYAEYHNIGPGSDKSKRVKWAGIWNLNSKAARWF 375
Query: 561 VPIRFFHGDDWIRVTRVPYSPGATK 585
P +FFHG DWI VT +P PG K
Sbjct: 376 SPSKFFHGTDWIEVTGIPCFPGVPK 450
>TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (38%)
Length = 678
Score = 199 bits (507), Expect = 2e-51
Identities = 96/199 (48%), Positives = 125/199 (62%), Gaps = 1/199 (0%)
Frame = +1
Query: 384 DRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQ 443
D S+FYKC +GYQDTLY H+ RQFYR+C I GT+DF+FG++ VLQNC R P N+
Sbjct: 1 DLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNP-PNK 177
Query: 444 QCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIG 503
+TAQGR + NQ TG+ I + A PV+ + YL RPWK +SRT+F+ TY+
Sbjct: 178 VNTITAQGRTDPNQNTGISIHNSRVTAASDLRPVQNSVRTYLGRPWKQYSRTVFMKTYLD 357
Query: 504 DMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAAS-FVP 562
+I P G+M W +G DT YYGEY N GPGS +RVKW G + ITS AS F
Sbjct: 358 GLINPAGWMEW---SGNFALDTLYYGEYMNTGPGSSTARRVKWSGYRVITSASEASKFSV 528
Query: 563 IRFFHGDDWIRVTRVPYSP 581
F G+ W+ T+VP++P
Sbjct: 529 ANFIAGNAWLPSTKVPFTP 585
>TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (39%)
Length = 601
Score = 190 bits (483), Expect = 1e-48
Identities = 95/197 (48%), Positives = 120/197 (60%)
Frame = +2
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
I F N+AG HQAVALR SD S+FY+C +GYQDTLY ++ RQFYR+C I GT+DF+F
Sbjct: 8 ITFRNTAGATNHQAVALRSGSDLSVFYRCSFEGYQDTLYVYSDRQFYRECDIYGTVDFIF 187
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
G++ V QNC R P N+ +TAQGR + NQ TG+ I + A VR
Sbjct: 188 GNAAVVFQNCNIYARNP-PNKVNTITAQGRTDPNQNTGISIHNSKVTAASDLMGVR---- 352
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
YL RPW+ +SRT+F+ TY+ +I PEG++ W +G T YYGEY N GPGS
Sbjct: 353 TYLGRPWQQYSRTVFMKTYLDSLINPEGWLEW---SGNFALSTLYYGEYMNTGPGSSTAN 523
Query: 543 RVKWQGVKTITSEGAAS 559
RV W G ITS AS
Sbjct: 524 RVNWLGYHVITSASEAS 574
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,015,618
Number of Sequences: 63676
Number of extensions: 293735
Number of successful extensions: 1398
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 1315
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1324
length of query: 593
length of database: 12,639,632
effective HSP length: 103
effective length of query: 490
effective length of database: 6,081,004
effective search space: 2979691960
effective search space used: 2979691960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144541.15


