

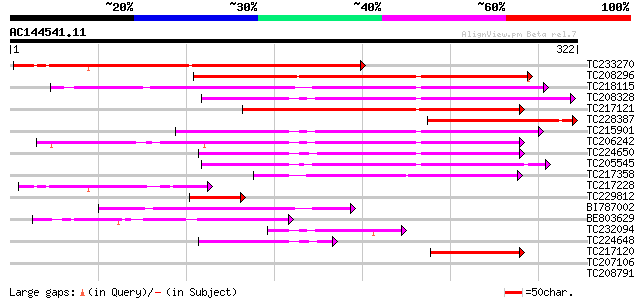
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.11 - phase: 0
(322 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC233270 similar to UP|O04018 (O04018) F7G19.1 protein, partial ... 271 4e-73
TC208296 similar to UP|O23548 (O23548) CLP proteinase like prote... 162 2e-40
TC218115 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp proteas... 125 2e-29
TC208328 similar to GB|AAO63325.1|28950803|BT005261 At5g23140 {A... 125 3e-29
TC217121 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)... 120 8e-28
TC228387 similar to UP|O04018 (O04018) F7G19.1 protein, partial ... 114 5e-26
TC215901 similar to UP|O82421 (O82421) Clp protease (Fragment) ,... 107 7e-24
TC206242 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 104 5e-23
TC224650 homologue to UP|O49081 (O49081) Clp-like energy-depende... 100 1e-21
TC205545 similar to PIR|T52454|T52454 ATP-dependent Clp proteina... 78 5e-15
TC217358 homologue to UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp p... 72 4e-13
TC217228 similar to UP|Q8H0P8 (Q8H0P8) RNA-binding protein, part... 67 8e-12
TC229812 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)... 56 3e-08
BI787002 similar to GP|14596089|gb| ATP-dependent Clp protease-l... 55 4e-08
BE803629 55 6e-08
TC232094 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 49 2e-06
TC224648 homologue to UP|O49081 (O49081) Clp-like energy-depende... 43 2e-04
TC217120 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)... 41 8e-04
TC207106 UP|Q70MR6 (Q70MR6) Ornithine decarboxylase , complete 37 0.016
TC208791 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 37 0.016
>TC233270 similar to UP|O04018 (O04018) F7G19.1 protein, partial (50%)
Length = 601
Score = 271 bits (692), Expect = 4e-73
Identities = 141/203 (69%), Positives = 167/203 (81%), Gaps = 3/203 (1%)
Frame = +1
Query: 3 TCFRVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPF---NPNDPFLSKLASVAES 59
T RVP S P + P S T T T+ R L+T +++ +KIP NP DPFLSKLAS+A S
Sbjct: 1 TSLRVPTSTPIT-PQSPTA-TRTKCRNLKTSCSVNAAKIPMPPLNPKDPFLSKLASIAAS 174
Query: 60 SPETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGR 119
SP+TL N + D P+LDIFDSP+LMATPAQ+ERS SY++ R P+RPPPDLPSLLL+GR
Sbjct: 175 SPDTLLNTPKNSDTPPYLDIFDSPKLMATPAQVERSVSYNEHR-PRRPPPDLPSLLLHGR 351
Query: 120 IVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYD 179
IVYIGMPLVPAVTELV+AELM+LQ+M PKEPIYIYINSTGTTR DGETV ME+EGFAIYD
Sbjct: 352 IVYIGMPLVPAVTELVVAELMYLQYMDPKEPIYIYINSTGTTRDDGETVGMETEGFAIYD 531
Query: 180 AMMQMKTEINTVALGAAVGQACL 202
AMMQ+K EI+TVA+G+A+GQACL
Sbjct: 532 AMMQLKNEIHTVAVGSAIGQACL 600
>TC208296 similar to UP|O23548 (O23548) CLP proteinase like protein, partial
(74%)
Length = 1003
Score = 162 bits (410), Expect = 2e-40
Identities = 91/195 (46%), Positives = 125/195 (63%), Gaps = 2/195 (1%)
Frame = +3
Query: 105 KRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRAD 164
++PPPDL S L RIVY+GM LVP+VTEL++AE ++LQ+ +PIY+YINSTGTT+
Sbjct: 129 EQPPPDLASYLYKNRIVYLGMSLVPSVTELILAEFLYLQYEDDDKPIYLYINSTGTTKG- 305
Query: 165 GETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQ 224
GE + E+E FAIYD M +K I T+ +G A G+A LLL+AG KG R P + MI+Q
Sbjct: 306 GEKLGYETEAFAIYDVMRYVKPPIFTLCVGNAWGEAALLLAAGAKGNRSALPSSTIMIRQ 485
Query: 225 PRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKE 284
P G A+DV + +EV + LVKL AKH E + E + ++R Y A E
Sbjct: 486 PIARFQG--QATDVNLARREVNNVKTELVKLYAKHMEKTPEQIEADIQRPKYFSPSEAVE 659
Query: 285 FGVIDKILW--RGQE 297
+G+IDK+++ RG E
Sbjct: 660 YGIIDKVIYNDRGSE 704
>TC218115 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp protease-like
protein (At5g45390), partial (77%)
Length = 1087
Score = 125 bits (315), Expect = 2e-29
Identities = 90/285 (31%), Positives = 145/285 (50%), Gaps = 2/285 (0%)
Frame = +3
Query: 24 PTRLRTLRTVNAISKSKIPFNPNDPFLSKLAS-VAESSPETLFNPSSS-PDNLPFLDIFD 81
P+ +RTL S K P + + F+ K S +++ P +F P+S P P F
Sbjct: 69 PSSIRTLS-----SNPKPPISSSFFFIPKPHSRFSKTPPRCVFTPASPIPSKNPN---FG 224
Query: 82 SPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMF 141
S T E S+ + + D+ LLL RIV++G + V + ++++++
Sbjct: 225 SQSRNPTSLTFELSSPQTPSTAARGAEGDVMGLLLRERIVFLGSSIDDFVADAIMSQMLL 404
Query: 142 LQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQAC 201
L P + I ++INSTG + S AIYDA+ ++ +++TVALG A A
Sbjct: 405 LDAQDPTKDIRLFINSTGGSL---------SATMAIYDAVQLVRADVSTVALGIAASTAS 557
Query: 202 LLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTE 261
++L GTKG+R+ P+ + MI QP +SG A DV I AKEVM N++ + ++++ T
Sbjct: 558 VILGGGTKGKRFAMPNTRIMIHQPLGGASG--QAIDVEIQAKEVMHNKNNITRIISSFTG 731
Query: 262 NSEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQEKIMADVPSR 306
S E V + R YM + A E+G+ID ++ R + VP R
Sbjct: 732 RSFEQVQKDIDRDKYMSPIEAVEYGIIDGVIDRDSIIPLMPVPER 866
>TC208328 similar to GB|AAO63325.1|28950803|BT005261 At5g23140 {Arabidopsis
thaliana;} , partial (88%)
Length = 1033
Score = 125 bits (313), Expect = 3e-29
Identities = 75/212 (35%), Positives = 116/212 (54%)
Frame = +2
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RI+ I P+ +V+A+L+FL+ P +PI +Y+NS G G A
Sbjct: 173 DIFSRLLKERIICINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG-----GAVTA 337
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M +++ +NT+ +G A LLL+AG KG R P+A MI QP
Sbjct: 338 ----GLAIYDTMQYIRSPVNTICMGQAASMGSLLLAAGAKGERRSLPNATIMIHQPSGGY 505
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
SG A D+ IH K ++ D+L +L AKHT S E + M R +M AKEFG+ID
Sbjct: 506 SG--QAKDIAIHTKHIVRVWDSLNELYAKHTGQSVEVIQTNMDRDNFMTPKEAKEFGLID 679
Query: 290 KILWRGQEKIMADVPSRDDRENGTAGAKVENR 321
+++ + +++D + ++ G+ ++E +
Sbjct: 680 EVIDQRPMALVSDAVGNEGKDKGSN*NQMEGK 775
>TC217121 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)
(At1g49970/F2J10_5), partial (42%)
Length = 988
Score = 120 bits (301), Expect = 8e-28
Identities = 64/160 (40%), Positives = 99/160 (61%)
Frame = +2
Query: 133 ELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVA 192
EL++A+ M+L + P +PIY+YINS+GT ETV E+E ++I D M +K ++ TV
Sbjct: 2 ELIVAQFMWLDYDNPSKPIYLYINSSGTLNEKNETVGSETEAYSIADMMSYVKADVYTVN 181
Query: 193 LGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTL 252
G A GQA +LLS GTKG R + P++ + P+V S D+ I AKE+ N +
Sbjct: 182 CGMAFGQAAMLLSLGTKGYRAVQPNSSTKLYLPKVNRSS-GAVIDMWIKAKELEANTEYY 358
Query: 253 VKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKIL 292
++LLAK T S+E ++ ++R Y+ A A ++G+ DKI+
Sbjct: 359 IELLAKGTGKSKEEIAKDVQRPKYLQAQDAIDYGIADKII 478
>TC228387 similar to UP|O04018 (O04018) F7G19.1 protein, partial (16%)
Length = 800
Score = 114 bits (286), Expect = 5e-26
Identities = 58/85 (68%), Positives = 68/85 (79%)
Frame = +3
Query: 238 VLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQE 297
+L +V++NRD LVKLLAKHT NSEETV+NVMKR YYMDA AKEFGVID+ILWRGQE
Sbjct: 144 ILFFFPQVIINRDNLVKLLAKHTGNSEETVANVMKRPYYMDATRAKEFGVIDRILWRGQE 323
Query: 298 KIMADVPSRDDRENGTAGAKVENRF 322
K+MADV S DD + G AG K+ + F
Sbjct: 324 KVMADVASPDDWDKG-AGIKIVDGF 395
>TC215901 similar to UP|O82421 (O82421) Clp protease (Fragment) , partial
(75%)
Length = 1214
Score = 107 bits (267), Expect = 7e-24
Identities = 64/209 (30%), Positives = 114/209 (53%)
Frame = +1
Query: 95 SASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIY 154
SAS + P+ D ++LL RI+++G + + +I++L+FL K+ I ++
Sbjct: 232 SASTASPWLPRFEELDATNMLLRQRIIFLGSQVDDMTADFIISQLLFLDAEDSKKDIKLF 411
Query: 155 INSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYM 214
INS G G A G IYDAM K +++TV LG A +L++GTKG+RY
Sbjct: 412 INSPG-----GSVTA----GMGIYDAMKLCKADVSTVCLGLAASMGAFILASGTKGKRYC 564
Query: 215 TPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRS 274
P+++ MI QP + G A+++ I +E+ ++ + K+L++ T EE + R
Sbjct: 565 MPNSRVMIHQPLGTAGG--KATEMSIRIREMAYHKIKINKILSRITGKPEEQIELDTDRD 738
Query: 275 YYMDALLAKEFGVIDKILWRGQEKIMADV 303
+M+ AKE+G++D ++ G+ ++A +
Sbjct: 739 NFMNPWEAKEYGLVDGVIDDGKPGLVAPI 825
>TC206242 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (86%)
Length = 1276
Score = 104 bits (260), Expect = 5e-23
Identities = 84/293 (28%), Positives = 146/293 (49%), Gaps = 16/293 (5%)
Frame = +1
Query: 16 PASSTRF-------TPTRLRTLRTVNAISKSK-IPFNPNDPFLSKLASVAESSPETLFNP 67
P+SS RF TP+ +V+ + S+ I + D ++K ++V + + P
Sbjct: 136 PSSSLRFNSLLVSSTPSSSSRSNSVSFPALSRNIRKSVEDRKINKNSAVKAVYGDEFWTP 315
Query: 68 S-SSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPP-------DLPSLLLNGR 119
+ SSP DI+ + + Q+ S + + + PPP + S L R
Sbjct: 316 ARSSPQ-----DIWS----IRSDLQVPSSPYFPTYAQGQGPPPMVAERFQSVISQLFQYR 468
Query: 120 IVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYD 179
I+ G + + +++A+L++L + P + I +Y+NS G G A G AI+D
Sbjct: 469 IIRCGGAVDDDMANIIVAQLLYLDAVDPNKDIVMYVNSPG-----GSVTA----GMAIFD 621
Query: 180 AMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVL 239
M ++ +++TV +G A LLSAGTKG+RY P+++ MI QP + G +D+
Sbjct: 622 TMRHIRPDVSTVCVGLAASMGAFLLSAGTKGKRYSLPNSRIMIHQPLGGAQG--GQTDID 795
Query: 240 IHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKIL 292
I A E++ ++ L LA HT S + ++ R ++M A AKE+G+ID ++
Sbjct: 796 IQANEMLHHKANLNGYLAYHTGQSLDKINQDTDRDFFMSAKEAKEYGLIDGVI 954
>TC224650 homologue to UP|O49081 (O49081) Clp-like energy-dependent protease,
partial (72%)
Length = 1067
Score = 100 bits (248), Expect = 1e-21
Identities = 64/185 (34%), Positives = 93/185 (49%)
Frame = +1
Query: 108 PPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGET 167
P DL S+L RI++IG P+ V + VI++L+ L + I +YIN G G T
Sbjct: 298 PLDLTSVLFRNRIIFIGQPVNSQVAQRVISQLVTLATINENADILVYINCPG-----GST 462
Query: 168 VAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRV 227
++ AIYD M +K ++ TV G A Q LLL+ G KG RY P+A+ MI QP+
Sbjct: 463 YSV----LAIYDCMSWIKPKVGTVCFGVAASQGALLLAGGEKGMRYAMPNARIMIHQPQS 630
Query: 228 PSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGV 287
G DV E + +R + K+ + T E V +R ++ A EFG+
Sbjct: 631 GCGG--HVEDVRRQVNEAVQSRHKIDKMYSVFTGQPLEKVQQYTERDRFLSVSEALEFGL 804
Query: 288 IDKIL 292
ID +L
Sbjct: 805 IDGVL 819
>TC205545 similar to PIR|T52454|T52454 ATP-dependent Clp proteinase
catalytic chain P2 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (81%)
Length = 1369
Score = 78.2 bits (191), Expect = 5e-15
Identities = 54/198 (27%), Positives = 97/198 (48%)
Frame = +3
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ + L R+++IG + + ++A +++L + + +Y+YIN G G+
Sbjct: 390 DVWNALYRERVIFIGQEIDEEFSNQILATMLYLDSIENSKKLYMYINGPG-----GDL-- 548
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ AIYD M +++ + T +G A A LL+AG K R P ++ + P +
Sbjct: 549 --TPSMAIYDTMQSLQSPVATHCVGYAYSLAAFLLAAGEKSNRSAMPLSRVALTSPAGAA 722
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A D+ A E++ RD L L+K T E ++ + R +A A E+G+ID
Sbjct: 723 RG--QADDIQNEANELLRIRDYLFNELSKKTGQPLEKITKDLSRMKRFNAQEALEYGLID 896
Query: 290 KILWRGQEKIMADVPSRD 307
+I+ +I AD P ++
Sbjct: 897 RIV--RPPRIKADAPRKE 944
>TC217358 homologue to UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease
proteolytic subunit (Endopeptidase Clp) , partial (76%)
Length = 1519
Score = 71.6 bits (174), Expect = 4e-13
Identities = 41/153 (26%), Positives = 75/153 (48%)
Frame = +1
Query: 139 LMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVG 198
+++L + +Y++INS G G AIYD M ++ ++ TV +G A
Sbjct: 19 MVYLSIEEENKDLYLFINSPG---------GWVIPGIAIYDTMQFVQPDVQTVCMGLAAS 171
Query: 199 QACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAK 258
LL+ G +R PHA+ MI QP S + ++ A+E++ R+T+ ++ +
Sbjct: 172 MGSFLLAGGEITKRLAFPHARVMIHQP-ASSFYEAQTGEFILEAEELLKMRETITRVYVQ 348
Query: 259 HTENSEETVSNVMKRSYYMDALLAKEFGVIDKI 291
T +S M+R +M A A+ +G++D +
Sbjct: 349 RTGKPLWVISEDMERDVFMSAAEAQAYGIVDLV 447
>TC217228 similar to UP|Q8H0P8 (Q8H0P8) RNA-binding protein, partial (36%)
Length = 698
Score = 67.4 bits (163), Expect = 8e-12
Identities = 48/113 (42%), Positives = 61/113 (53%), Gaps = 3/113 (2%)
Frame = +1
Query: 6 RVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPF---NPNDPFLSKLASVAESSPE 62
RVP S P + P S T T T+ R L+T +++ +KIP NP DPFLSKLAS+A SSP+
Sbjct: 10 RVPTSTPIT-PQSPTA-TRTKCRNLKTSCSVNAAKIPMPPLNPKDPFLSKLASIAASSPD 183
Query: 63 TLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLL 115
TL N + D PFLD T A+ ++ S R PPD S L
Sbjct: 184 TLLNTPKNSDTPPFLD-------RGTNAK-DKMPGLSSGRTDHSLPPDATSTL 318
>TC229812 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)
(At1g49970/F2J10_5), partial (34%)
Length = 558
Score = 55.8 bits (133), Expect = 3e-08
Identities = 25/32 (78%), Positives = 28/32 (87%)
Frame = +1
Query: 103 RPKRPPPDLPSLLLNGRIVYIGMPLVPAVTEL 134
RPK PPDLPSLLL+ RI Y+GMP+VPAVTEL
Sbjct: 463 RPKTAPPDLPSLLLDARICYLGMPIVPAVTEL 558
>BI787002 similar to GP|14596089|gb| ATP-dependent Clp protease-like protein
{Arabidopsis thaliana}, partial (31%)
Length = 438
Score = 55.1 bits (131), Expect = 4e-08
Identities = 39/146 (26%), Positives = 69/146 (46%)
Frame = +3
Query: 51 SKLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPD 110
S+ + +++ +F S P +P Q E SA + + D
Sbjct: 36 SRFSKPSKTPARCVFTASPIPSKIPNF----GSQSRNPGLTFELSAPQTPSTAARGAEGD 203
Query: 111 LPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAM 170
+ LLL RIV++G + V + ++++L+ L + P + I ++INSTG +
Sbjct: 204 VMVLLLRERIVFLGSSIDDFVADAIMSQLLLLDALDPTKDIRLFINSTGGSL-------- 359
Query: 171 ESEGFAIYDAMMQMKTEINTVALGAA 196
S AIYDA+ ++ +++TVALG A
Sbjct: 360 -SATMAIYDAVQLVRADVSTVALGIA 434
>BE803629
Length = 583
Score = 54.7 bits (130), Expect = 6e-08
Identities = 46/157 (29%), Positives = 75/157 (47%), Gaps = 9/157 (5%)
Frame = +2
Query: 14 SMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESS---------PETL 64
++P+SS TP +R +N S S + + + F++ A + +S P +L
Sbjct: 50 ALPSSSRTVTP-----IRAMNT-STSPVRTSLSTSFVAPYAGICSASSPFSGHKLRPSSL 211
Query: 65 FNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRIVYIG 124
NP+S FL ++ +R +++ Q PPPDL S L RIV +G
Sbjct: 212 -NPAS------FLGSNAKRGVVTMVIPFQRGSAWEQ------PPPDLASYLYKNRIVILG 352
Query: 125 MPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTT 161
M L P+ L++ E ++ + P+Y+YINST TT
Sbjct: 353 MSL*PSAAALILPEFLYR*YEHDD*PMYLYINSTVTT 463
>TC232094 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (49%)
Length = 842
Score = 49.3 bits (116), Expect = 2e-06
Identities = 31/99 (31%), Positives = 47/99 (47%), Gaps = 20/99 (20%)
Frame = +2
Query: 147 PKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLS- 205
P + I +Y++S G G A G AI+D M ++ +++TV +G A + LLS
Sbjct: 458 PNKDIVMYVSSPG-----GSVTA----GMAIFDTMRHIRPDVSTVCIGLAASKGAFLLSA 610
Query: 206 -------------------AGTKGRRYMTPHAKAMIQQP 225
AGTKG+RY P+++ MI QP
Sbjct: 611 GTKGNDSFTMSSKGAFLLSAGTKGKRYSLPNSRIMIHQP 727
>TC224648 homologue to UP|O49081 (O49081) Clp-like energy-dependent protease,
partial (34%)
Length = 678
Score = 43.1 bits (100), Expect = 2e-04
Identities = 27/79 (34%), Positives = 42/79 (52%)
Frame = +2
Query: 108 PPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGET 167
P DL S+L RI++IG P+ V + VI++L+ L + I +YIN G G T
Sbjct: 302 PLDLSSVLFRNRIIFIGQPVNSQVAQRVISQLVTLATIDENADILVYINCPG-----GST 466
Query: 168 VAMESEGFAIYDAMMQMKT 186
++ AIYD M +++
Sbjct: 467 YSV----LAIYDCMSWVRS 511
>TC217120 similar to UP|Q9XJ35 (Q9XJ35) NClpP5 (F2J10.14 protein)
(At1g49970/F2J10_5), partial (15%)
Length = 713
Score = 40.8 bits (94), Expect = 8e-04
Identities = 20/53 (37%), Positives = 34/53 (63%)
Frame = +3
Query: 240 IHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKIL 292
I AKE+ N + ++LLAK T S+E ++ ++R Y+ A A ++G+ DKI+
Sbjct: 33 IKAKELEANTEYYIELLAKGTGKSKEEIAKDVQRPKYLQAQDAIDYGIADKII 191
>TC207106 UP|Q70MR6 (Q70MR6) Ornithine decarboxylase , complete
Length = 1551
Score = 36.6 bits (83), Expect = 0.016
Identities = 26/69 (37%), Positives = 39/69 (55%), Gaps = 1/69 (1%)
Frame = +1
Query: 8 PISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNP-NDPFLSKLASVAESSPETLFN 66
P S P S P+S+T+ +P+ +R+LR+ A S P P ++PF + ESS +T N
Sbjct: 247 PRSSP-STPSSATQTSPSSVRSLRSAPA---STAPAKPRSNPFSPSESHPTESSMQTHAN 414
Query: 67 PSSSPDNLP 75
PS + D P
Sbjct: 415 PSHTSDTPP 441
>TC208791 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (14%)
Length = 618
Score = 36.6 bits (83), Expect = 0.016
Identities = 17/39 (43%), Positives = 25/39 (63%)
Frame = +2
Query: 202 LLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLI 240
+LLSAGTKG RY P+++ MI QP + G + D+ +
Sbjct: 128 ILLSAGTKGNRYSLPNSRIMIHQPLGGAQGGQTDIDIQV 244
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,280,560
Number of Sequences: 63676
Number of extensions: 154697
Number of successful extensions: 1264
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 1232
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1251
length of query: 322
length of database: 12,639,632
effective HSP length: 97
effective length of query: 225
effective length of database: 6,463,060
effective search space: 1454188500
effective search space used: 1454188500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144541.11


