

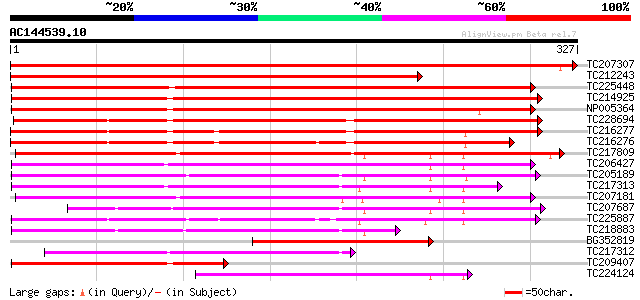
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144539.10 - phase: 0
(327 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207307 homologue to UP|E13B_PHAVU (P23535) Glucan endo-1,3-bet... 517 e-147
TC212243 UP|O49011 (O49011) Beta-1,3-glucanase 1 (Fragment) , co... 354 3e-98
TC225448 similar to UP|Q39900 (Q39900) Beta-1,3-glucanase , part... 348 2e-96
TC214925 UP|E13A_SOYBN (Q03773) Glucan endo-1,3-beta-glucosidase... 342 1e-94
NP005364 beta-1,3-glucanase 340 7e-94
TC228694 UP|Q6S9W0 (Q6S9W0) Endo-1,3-beta-glucanase, complete 322 2e-88
TC216277 UP|O49012 (O49012) Beta-1,3-glucanase 3 (Fragment) , co... 278 2e-75
TC216276 homologue to UP|E13B_SOYBN (P52395) Glucan endo-1,3-bet... 268 3e-72
TC217809 beta-1,3-glucanase 7 225 2e-59
TC206427 similar to UP|Q8S9I6 (Q8S9I6) AT5g55180/MCO15_13, parti... 201 4e-52
TC205189 similar to UP|Q94G86 (Q94G86) Beta-1,3-glucanase-like p... 197 5e-51
TC217313 weakly similar to UP|Q9LVZ9 (Q9LVZ9) Beta-1,3-glucanase... 194 3e-50
TC207181 183 1e-46
TC207687 178 3e-45
TC225887 similar to UP|Q9M4A9 (Q9M4A9) Beta-1,3 glucanase precur... 176 1e-44
TC218883 weakly similar to UP|Q9MAQ2 (Q9MAQ2) CDS, partial (54%) 151 4e-37
BG352819 homologue to SP|Q03773|E13A Glucan endo-1 3-beta-glucos... 130 8e-31
TC217312 similar to GB|AAO63352.1|28950857|BT005288 At2g26600 {A... 129 2e-30
TC209407 UP|E13A_SOYBN (Q03773) Glucan endo-1,3-beta-glucosidase... 123 1e-28
TC224124 similar to UP|Q8L9D9 (Q8L9D9) Beta-1,3-glucanase-like p... 112 2e-25
>TC207307 homologue to UP|E13B_PHAVU (P23535) Glucan
endo-1,3-beta-glucosidase, basic isoform
((1->3)-beta-glucan endohydrolase)
((1->3)-beta-glucanase) (Beta-1,3-endoglucanase) ,
partial (91%)
Length = 1539
Score = 517 bits (1332), Expect = e-147
Identities = 253/331 (76%), Positives = 286/331 (85%), Gaps = 4/331 (1%)
Frame = +2
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M+GNNLPS + I L +SNNIKRMRLYDPNQAALEALRNSGIEL+LGVPNSDLQ +ATN
Sbjct: 362 MLGNNLPSANDVIGLYRSNNIKRMRLYDPNQAALEALRNSGIELILGVPNSDLQGLATNP 541
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
D + QWVQKNVLNF+PSVKIKY+AVGNEV+PVGGSS A++VLPAIQN+YQAIRA+ LHD
Sbjct: 542 DTSRQWVQKNVLNFWPSVKIKYVAVGNEVSPVGGSSSVAQYVLPAIQNVYQAIRAQGLHD 721
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVST+IDMT+IG S+PPS+GSFR DVRSYLDPIIGYLVYANAPL N+Y YFSY NP
Sbjct: 722 QIKVSTSIDMTLIGNSFPPSQGSFRGDVRSYLDPIIGYLVYANAPLLVNVYPYFSYTGNP 901
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFT+PNVVVWDG GYQNLFDA+LDS+HAAIDNT IG+V+VVVSESGWPSDG
Sbjct: 902 RDISLPYALFTAPNVVVWDGQYGYQNLFDAMLDSVHAAIDNTKIGYVEVVVSESGWPSDG 1081
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMR-SGPIETYIFGLFDENQKNPELEKHFGVFYPNK 299
GFA TYDNARVYLDNL+R G+P R S P ETYIF +FDENQKNPE+EKHFG+F PNK
Sbjct: 1082GFAATYDNARVYLDNLVRRANRGSPRRPSKPTETYIFAMFDENQKNPEIEKHFGLFNPNK 1261
Query: 300 QKKYPFGFQGKIDGTNL---FNVSFPLKSDI 327
QKKYPFGF GK G + FN + +KSD+
Sbjct: 1262QKKYPFGFGGKRLGKVVIDDFNATTSIKSDV 1354
>TC212243 UP|O49011 (O49011) Beta-1,3-glucanase 1 (Fragment) , complete
Length = 741
Score = 354 bits (909), Expect = 3e-98
Identities = 171/238 (71%), Positives = 202/238 (84%)
Frame = +3
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMG+NLP E + L KSN+I RMR+Y+P+QAAL+AL NSGIEL+LGV + DLQ +ATN
Sbjct: 24 MMGDNLPPANEVVSLYKSNDIMRMRIYNPDQAALQALGNSGIELILGVLHQDLQGLATNA 203
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
A QWVQ NVLNF+PSVKIK++ VGNE+NPVG SS+FA++VLPAIQNIYQAIRA+ L D
Sbjct: 204 STAQQWVQSNVLNFWPSVKIKHVVVGNEINPVGSSSEFAQYVLPAIQNIYQAIRAQGLQD 383
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKV+TAIDMT++G SYPPS+ FR+DVRSYLDPIIGYLVYANAPL AN+ YFSY +NP
Sbjct: 384 LIKVTTAIDMTLLGNSYPPSQSYFRTDVRSYLDPIIGYLVYANAPLLANVLPYFSYSNNP 563
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPS 238
DISL YALF S NVVVWDG GYQNLFDA+LD++H AIDNTGIG+V+VVVSE GWP+
Sbjct: 564 IDISLSYALFNSTNVVVWDGQYGYQNLFDAMLDAVHVAIDNTGIGYVEVVVSERGWPN 737
>TC225448 similar to UP|Q39900 (Q39900) Beta-1,3-glucanase , partial (93%)
Length = 1098
Score = 348 bits (892), Expect = 2e-96
Identities = 174/304 (57%), Positives = 230/304 (75%), Gaps = 2/304 (0%)
Frame = +2
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+GNNLPS +E + L K + +RMR+YDP+Q L+ALR S IEL+L +PN +LQ++A++ D
Sbjct: 65 VGNNLPSPQEVVALYKQYDFRRMRIYDPSQQVLQALRGSNIELLLDLPNVNLQSVASSQD 244
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A +WVQ NV N+ +V+ +YI+VGNEV P FA+FV+PAIQNI +A+ A L +Q
Sbjct: 245 NANRWVQDNVRNYANNVRFRYISVGNEVKPW---DSFARFVVPAIQNIQRAVSAAGLGNQ 415
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSD-VRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVSTAI+ + SYPPS+GSFRSD + SYLD +I +LV NAPL N+Y YF+Y NP
Sbjct: 416 IKVSTAIETGALAESYPPSRGSFRSDYLTSYLDGVIRHLVNNNAPLLVNVYPYFAYIGNP 595
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALF SP+VVV DGS GY+NLF+A++D+++AA++ G G + +VVSESGWPS G
Sbjct: 596 RDISLDYALFRSPSVVVQDGSLGYRNLFNAMVDAVYAALEKAGGGSLNIVVSESGWPSSG 775
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSG-PIETYIFGLFDENQKNPELEKHFGVFYPNK 299
G AT+ DNAR Y NL+R+VK GTP R G P+ETY+F +F+ENQK PE EK +G+F PNK
Sbjct: 776 GTATSLDNARTYNTNLVRNVKQGTPKRPGRPLETYVFAMFEENQKQPEYEKFWGLFLPNK 955
Query: 300 QKKY 303
Q KY
Sbjct: 956 QLKY 967
>TC214925 UP|E13A_SOYBN (Q03773) Glucan endo-1,3-beta-glucosidase precursor
((1->3)-beta-glucan endohydrolase)
((1->3)-beta-glucanase) (Beta-1,3-endoglucanase) ,
complete
Length = 1299
Score = 342 bits (877), Expect = 1e-94
Identities = 171/309 (55%), Positives = 227/309 (73%), Gaps = 3/309 (0%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+GNNLP+ +E + L NI+RMR+Y P+ LEALR S IEL+L +PN +L+N+A++ D
Sbjct: 190 LGNNLPTPQEVVALYNQANIRRMRIYGPSPEVLEALRGSNIELLLDIPNDNLRNLASSQD 369
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A +WVQ N+ N+ +V+ +Y++VGNEV P FA+F++PA++NI +AI L +Q
Sbjct: 370 NANKWVQDNIKNYANNVRFRYVSVGNEVKP---EHSFAQFLVPALENIQRAISNAGLGNQ 540
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSDVR-SYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
+KVSTAID + S+PPSKGSF+SD R +YLD +I +LV NAPL N+YSYF+Y NP
Sbjct: 541 VKVSTAIDTGALAESFPPSKGSFKSDYRGAYLDGVIRFLVNNNAPLMVNVYSYFAYTANP 720
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
KDISL YALF SP+VVV DGS GY+NLFDA +D+++AA++ G G + +VVSESGWPS G
Sbjct: 721 KDISLDYALFRSPSVVVQDGSLGYRNLFDASVDAVYAALEKAGGGSLNIVVSESGWPSSG 900
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSG-PIETYIFGLFDENQKNPELEKHFGVFYP-N 298
G AT+ DNAR Y NL+R+VK GTP R G P+ETY+F +FDENQK PE EK +G+F P
Sbjct: 901 GTATSLDNARTYNTNLVRNVKQGTPKRPGAPLETYVFAMFDENQKQPEFEKFWGLFSPIT 1080
Query: 299 KQKKYPFGF 307
KQ KY F
Sbjct: 1081KQPKYSINF 1107
>NP005364 beta-1,3-glucanase
Length = 1047
Score = 340 bits (871), Expect = 7e-94
Identities = 170/305 (55%), Positives = 225/305 (73%), Gaps = 3/305 (0%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+GNNLPS +E + L K + +RMR+YDP+Q LEALR S IEL+L +PN +LQN+A + D
Sbjct: 124 IGNNLPSPQEVVALFKQYDFRRMRIYDPSQEVLEALRGSNIELLLDIPNDNLQNLAFSQD 303
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A +W+Q N+ N+ +V+ +YI+VGNEV P FA+F++PA+QNI +AI L +Q
Sbjct: 304 NANKWLQDNIKNYANNVRFRYISVGNEVKP---EHSFAQFLVPAMQNIQRAISNAGLGNQ 474
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSDVRS-YLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVSTAI+ + SYPPS GSFRSD R+ YLD +I +LV N PL N+Y YF+Y ++P
Sbjct: 475 IKVSTAIETGALADSYPPSMGSFRSDYRTAYLDGVIRHLVNNNTPLLVNVYPYFAYINDP 654
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
++ISL YALF SP+VVV DGS GY+NLFDA++D+++AA++ G G V +VVSESGWPS G
Sbjct: 655 RNISLDYALFRSPSVVVQDGSLGYRNLFDAMVDAVYAALEKAGGGSVSIVVSESGWPSSG 834
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSG--PIETYIFGLFDENQKNPELEKHFGVFYPN 298
G AT+ DNAR Y NL+R+VK GTP R P+ETY+F +F+EN K PE EK +GVF PN
Sbjct: 835 GTATSLDNARTYNTNLVRNVKQGTPKRPAGRPLETYVFAMFNENHKQPEYEKFWGVFLPN 1014
Query: 299 KQKKY 303
KQ KY
Sbjct: 1015KQPKY 1029
>TC228694 UP|Q6S9W0 (Q6S9W0) Endo-1,3-beta-glucanase, complete
Length = 1251
Score = 322 bits (824), Expect = 2e-88
Identities = 168/307 (54%), Positives = 210/307 (67%), Gaps = 2/307 (0%)
Frame = +3
Query: 3 GNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDI 62
GNNLP+++ +DL KSN I ++RLY P++ L+ALR S IE++LGVPN LQ++ TN
Sbjct: 180 GNNLPTKQAVVDLYKSNRIGKIRLYYPDEGVLQALRGSNIEVILGVPNDQLQSL-TNAGA 356
Query: 63 AIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQI 122
A WV K V + +VK KYIAVGNE++P A VLPA++NI +AI A NL Q+
Sbjct: 357 ATNWVNKYVKAYSQNVKFKYIAVGNEIHP---GDSLAGSVLPALENIQKAISAANLQGQM 527
Query: 123 KVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPKD 182
KVSTAID T++G SYPP G F S SY+ PI+ +L APL AN+Y YF+Y +N +
Sbjct: 528 KVSTAIDTTLLGNSYPPKDGVFSSSASSYIRPIVNFLARNGAPLLANVYPYFAYVNNQQS 707
Query: 183 ISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGGF 242
I L YALFT + GYQNLFDALLDSL+AA++ G VKVVVSESGWPS+GG
Sbjct: 708 IGLDYALFTKHG----NNEVGYQNLFDALLDSLYAALEKVGAPNVKVVVSESGWPSEGGV 875
Query: 243 ATTYDNARVYLDNLIRHVKGGTPMR-SGPIETYIFGLFDENQKN-PELEKHFGVFYPNKQ 300
T NA Y NLI H KGGTP R SGPIETY+F +FDENQK+ PE+E+HFG+F P+K
Sbjct: 876 GATVQNAGTYYRNLINHAKGGTPKRPSGPIETYLFAMFDENQKDGPEIERHFGLFRPDKS 1055
Query: 301 KKYPFGF 307
KY F
Sbjct: 1056PKYQLSF 1076
>TC216277 UP|O49012 (O49012) Beta-1,3-glucanase 3 (Fragment) , complete
Length = 1087
Score = 278 bits (711), Expect = 2e-75
Identities = 155/313 (49%), Positives = 204/313 (64%), Gaps = 6/313 (1%)
Frame = +3
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
++GNNLPS++E +DL K+N I RMR+Y P++ AL+ALR SGIEL++ V LQ++ T+
Sbjct: 24 VLGNNLPSRQEVVDLYKTNGIGRMRIYYPDEEALQALRGSGIELIMDVAKETLQSM-TDP 200
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WV K V + V KYIAVGNE++P ++ A+++L A+ NI AI + NL
Sbjct: 201 NAATDWVNKYVTAYSQDVNFKYIAVGNEIHP---NTNEAQYILSAMTNIQNAISSANL-- 365
Query: 121 QIKVSTAIDMTMIGT-SYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSY-KD 178
QIKVSTAID T I SYPP+ F SD Y+ PII +LV APL AN+Y YF+Y D
Sbjct: 366 QIKVSTAIDSTFIAPPSYPPNDAVFTSDAEPYVKPIIDFLVRNEAPLLANVYPYFAYAND 545
Query: 179 NPKDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPS 238
I L YALFT + GYQNLFDA+LDS++AA++ G +++VVSESGWPS
Sbjct: 546 QQNSIPLAYALFTQQG----NNDAGYQNLFDAMLDSIYAAVEKVGASNLQIVVSESGWPS 713
Query: 239 DGGFATTYDNARVYLDNLIRHVK--GGTPMRSG-PIETYIFGLFDENQK-NPELEKHFGV 294
+GG + DNA Y NLIRH GTP R G IETY+F +FDENQK + ++HFG+
Sbjct: 714 EGGAGASIDNAGTYYANLIRHASSGNGTPKRPGESIETYLFAMFDENQKQGADTDRHFGL 893
Query: 295 FYPNKQKKYPFGF 307
F P+K KY F
Sbjct: 894 FNPDKSPKYQLSF 932
>TC216276 homologue to UP|E13B_SOYBN (P52395) Glucan
endo-1,3-beta-glucosidase ((1->3)-beta-glucan
endohydrolase) ((1->3)-beta-glucanase)
(Beta-1,3-endoglucanase) (Fragment) , partial (95%)
Length = 1180
Score = 268 bits (684), Expect = 3e-72
Identities = 148/295 (50%), Positives = 198/295 (66%), Gaps = 4/295 (1%)
Frame = +3
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
++G+NLPS++E +DL K+N I RMR+Y P++ AL+ALR SGIEL++ V LQ++ T++
Sbjct: 117 VIGDNLPSRQEVVDLYKTNGIGRMRIYYPDEEALQALRGSGIELIMDVAKETLQSL-TDS 293
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WV K V + V KYIAVGNE++P ++ A+++L A+ NI AI + NL
Sbjct: 294 NAATDWVNKYVTPYSQDVNFKYIAVGNEIHP---NTNEAQYILSAMTNIQNAISSANL-- 458
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVSTAID T+I SYPP+ G F SD Y+ PII +LV APL AN+Y YF+Y N
Sbjct: 459 QIKVSTAIDSTLITNSYPPNDGVFTSDAEPYIKPIINFLVSNGAPLLANVYPYFAYA-ND 635
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+ I L YALFT + GYQNLFDA+LDS++AA++ G +++VVSESGWPS+G
Sbjct: 636 QSIPLAYALFTQQG----NNDVGYQNLFDAMLDSIYAALEKVGASNLQIVVSESGWPSEG 803
Query: 241 GFATTYDNARVYLDNLIRHVK--GGTPMRSG-PIETYIFGLFDENQK-NPELEKH 291
G + DNA Y NLIRH GTP R G IETY+F +FDENQK + E+H
Sbjct: 804 GAGASIDNAGTYYANLIRHASSGNGTPKRPGESIETYLFAMFDENQKQGADTERH 968
>TC217809 beta-1,3-glucanase 7
Length = 1867
Score = 225 bits (574), Expect = 2e-59
Identities = 127/328 (38%), Positives = 201/328 (60%), Gaps = 11/328 (3%)
Frame = +1
Query: 4 NNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIA 63
++LP+ + L + + IK +R+YD N L+A N+GIELM+GVPNSDL + + A
Sbjct: 283 DDLPTPDKVAQLVQLHKIKYVRIYDSNIQVLKAFANTGIELMIGVPNSDLLSFSQFQSNA 462
Query: 64 IQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQIK 123
W++ +VL +YP+ KI YI VG EV ++ + FV+PA+ N+ A++ LH +IK
Sbjct: 463 DSWLKNSVLPYYPATKIAYITVGAEVTESPNNA--SSFVVPAMTNVLTALKKLGLHKKIK 636
Query: 124 VSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPKDI 183
VS+ + ++ S+PPS G+F S +L P++ +L AP +IY Y++++D+ +
Sbjct: 637 VSSTHSLGVLSRSFPPSAGAFNSSHAHFLKPMLEFLAENQAPFMIDIYPYYAHRDSRSKV 816
Query: 184 SLQYALFTSPNVVVWDGSRG--YQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGG 241
SL YALF + + V+ D + G Y N+FDA +D+++ A+ +KV+V+E+GWPS G
Sbjct: 817 SLDYALFDASSEVI-DPNTGLLYTNMFDAQIDAIYFALMALNFRTIKVMVTETGWPSKGS 993
Query: 242 ---FATTYDNARVYLDNLIRHV--KGGTPMRSG-PIETYIFGLFDENQK-NPELEKHFGV 294
A T DNA+ Y NLIRHV GTP + G ++ YIF LF+EN+K E E+++G+
Sbjct: 994 PKETAATPDNAQTYNTNLIRHVINNTGTPAKPGEELDVYIFSLFNENRKPGLESERNWGL 1173
Query: 295 FYPNKQKKYPFGFQGK--IDGTNLFNVS 320
FYP++ Y F G+ +D T N++
Sbjct: 1174FYPDQTSVYSLDFTGRGAVDMTTEANIT 1257
>TC206427 similar to UP|Q8S9I6 (Q8S9I6) AT5g55180/MCO15_13, partial (89%)
Length = 1660
Score = 201 bits (511), Expect = 4e-52
Identities = 112/311 (36%), Positives = 183/311 (58%), Gaps = 9/311 (2%)
Frame = +3
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ N+LP+ + ++L KS + R++LYD + L A NSG+++++ +PN L N A
Sbjct: 111 IANDLPTPAKVVELLKSQGLNRVKLYDTDATVLTAFANSGMKVVVAMPNELLANAAAEQS 290
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
WVQ N+ ++YP+ +I+ IAVGNEV + KF++PA++N++ ++ +L
Sbjct: 291 FTDAWVQANISSYYPATQIEAIAVGNEV--FVDPNNTTKFLVPAMKNVHASLVKYSLDKN 464
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSD-VRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IK+S+ I ++ + S+P S GSF+++ + + P++ L + L N Y +F+Y N
Sbjct: 465 IKISSPIALSALQNSFPASSGSFKTELLEPVIKPMLDLLRQTGSYLMVNAYPFFAYAANS 644
Query: 181 KDISLQYALF-TSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
ISL YALF +P VV Y NLFDA +D++ AA+ VK+ VSE+GWPS
Sbjct: 645 DKISLDYALFKENPGVVDSGNGLKYTNLFDAQIDAVFAAMSALKYDDVKIAVSETGWPSA 824
Query: 240 GG---FATTYDNARVYLDNLIRHV--KGGTPMRSG-PIETYIFGLFDENQK-NPELEKHF 292
G + DNA Y NL++ V GTP++ ++ ++F LF+ENQK P E+++
Sbjct: 825 GDSNEIGASPDNAASYNGNLVKRVLSGSGTPLKQNESLDVFLFALFNENQKTGPTSERNY 1004
Query: 293 GVFYPNKQKKY 303
G+FYP ++K Y
Sbjct: 1005GLFYPTEKKVY 1037
>TC205189 similar to UP|Q94G86 (Q94G86) Beta-1,3-glucanase-like protein,
partial (91%)
Length = 1715
Score = 197 bits (501), Expect = 5e-51
Identities = 114/314 (36%), Positives = 182/314 (57%), Gaps = 9/314 (2%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ +NLP+ + +L KS I ++RLY + A ++AL NSGI +++G N D+ ++A + +
Sbjct: 226 VADNLPAPEDTANLLKSTTIGKVRLYGADPAIIKALANSGIGIVIGAANGDIPSLAADPN 405
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A QWV NVL +YP+ I I VGNE+ + ++ V PA++N+ A+ A +L +
Sbjct: 406 AATQWVNANVLPYYPASNITLITVGNEILTLADQGLKSQLV-PAMRNVQNALGAASLGGK 582
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPK 181
IKVST M ++ S PPS G F ++ L ++ L +P N Y +F+Y+ +P+
Sbjct: 583 IKVSTVHSMAVLTQSDPPSSGLFNPALQDTLKQLLALLKDNKSPFTINPYPFFAYQSDPR 762
Query: 182 DISLQYALFTSPNVVVWDGSRG--YQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
+L + LF PN D G Y N+FDA +D++H+A+ G V++VV+E+GWPS
Sbjct: 763 PETLAFCLF-QPNSGRVDSGNGKLYTNMFDAQVDAVHSALSAMGFQDVEIVVAETGWPSR 939
Query: 240 GG---FATTYDNARVYLDNLIRHVKG--GTPMRSG-PIETYIFGLFDENQK-NPELEKHF 292
G + +NA+ Y NLI H++ GTP+ G ++TYIF L+DE+ K P E+ F
Sbjct: 940 GDSNEVGPSVENAKAYNGNLIAHLRSLVGTPLMPGKSVDTYIFALYDEDLKPGPGSERAF 1119
Query: 293 GVFYPNKQKKYPFG 306
G+F ++ Y G
Sbjct: 1120GMFKTDRTVLYDVG 1161
>TC217313 weakly similar to UP|Q9LVZ9 (Q9LVZ9) Beta-1,3-glucanase-like
protein, partial (67%)
Length = 1142
Score = 194 bits (494), Expect = 3e-50
Identities = 103/292 (35%), Positives = 175/292 (59%), Gaps = 9/292 (3%)
Frame = +2
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ NN+PS E + L ++ I+ +R+YD + + L+A +G+E+++G+PN LQ++++N D
Sbjct: 257 IANNIPSPDEVVTLLRAAKIRNVRIYDADHSVLKAFSGTGLEIVVGLPNGQLQDMSSNPD 436
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSS-QFAKFVLPAIQNIYQAIRAKNLHD 120
A+ WV++NV +F P +I+ IAVGNEV +GG+ +L A++NIY A + +L
Sbjct: 437 HALNWVKENVQSFLPDTRIRGIAVGNEV--LGGTDYSLWGVLLGAVKNIYNATKKLHLDQ 610
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
+++STA + SYPPS G F ++V Y+ P++ + +P N Y + +Y +P
Sbjct: 611 LVQISTANSFAVFAVSYPPSSGKFDNNVNQYMKPLLEFFQQIGSPFCLNAYPFLAYAGDP 790
Query: 181 KDISLQYALFTSPNVVVWDG--SRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPS 238
+ I + YALF P ++D Y N+ DA +D+ +AA+++ G ++V+++E+GW S
Sbjct: 791 EHIDINYALF-EPTKGIYDPMYHLHYDNMLDAQIDAAYAALEDAGFDKMEVIITETGWAS 967
Query: 239 DGG---FATTYDNARVYLDNLIRHV--KGGTPMR-SGPIETYIFGLFDENQK 284
+G NAR Y NL R + + GTP R ++ YIF LF+EN+K
Sbjct: 968 NGDQTEAGANATNARTYNHNLRRRLAKRKGTPHRPKNVVKAYIFALFNENEK 1123
>TC207181
Length = 1691
Score = 183 bits (464), Expect = 1e-46
Identities = 109/311 (35%), Positives = 175/311 (56%), Gaps = 11/311 (3%)
Frame = +3
Query: 4 NNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIA 63
+NLPS + + + I +R+YD N L+ L + I +++ VPN+ L I ++N A
Sbjct: 12 SNLPSASDLVAFLQLQKITHVRIYDANSDILKTLSGTKIRVIISVPNNQLLAIGSSNSTA 191
Query: 64 IQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQIK 123
W+ +NV+ +YP I I+VG+EV SS A +LPA++++Y A+ A NLH QIK
Sbjct: 192 ASWIDRNVVAYYPQTLISGISVGDEVLTTVPSS--APLILPAVESLYNALVASNLHQQIK 365
Query: 124 VSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPKDI 183
VST ++I +PPS+ F + S + P++ +L +PL N+Y Y+ + N +
Sbjct: 366 VSTPHAASIILDPFPPSQAYFNQSLVSVILPLLQFLSRTGSPLMMNLYPYYVFMQNKGVV 545
Query: 184 SLQYALF--TSPNVVVWDGSR--GYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
L ALF +PN + D + Y N+ DA++D+ + ++ N I V V+V+E+GWP+
Sbjct: 546 PLDNALFKPLTPNKEMVDPNTLLHYTNVLDAMVDAAYFSMKNLNITDVAVLVTETGWPAK 725
Query: 240 GGFATTY---DNARVYLDNLIRHV--KGGTPMRSGPIET-YIFGLFDENQKNPEL-EKHF 292
G Y DNA Y NLIRHV + GTP+ + +I+ LF+E+ + P + E ++
Sbjct: 726 GDSKEPYATKDNADTYNSNLIRHVFDRTGTPLHPETTSSVFIYELFNEDLRAPPVSEANW 905
Query: 293 GVFYPNKQKKY 303
G+FY N Y
Sbjct: 906 GLFYGNTSPAY 938
>TC207687
Length = 1380
Score = 178 bits (452), Expect = 3e-45
Identities = 105/285 (36%), Positives = 160/285 (55%), Gaps = 9/285 (3%)
Frame = +1
Query: 34 LEALRNSGIELMLGVPNSDLQNIATNNDIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVG 93
L A NS +E ++G+ N LQ++ + A WVQ++V + +I I VGNEV
Sbjct: 25 LSAFSNSDVEFIIGLGNEYLQSMRDPSK-AQSWVQQHVQPYISQTRITCITVGNEVFNYN 201
Query: 94 GSSQFAKFVLPAIQNIYQAIRAKNLHDQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLD 153
+Q +LPA+Q++Y A+ L Q+ V+TA ++ S+PPS G+FR D+ Y+
Sbjct: 202 -DTQLTANLLPAMQSVYNALVNLGLAQQVTVTTAHSFNILANSFPPSSGAFRQDLIQYIQ 378
Query: 154 PIIGYLVYANAPLFANIYSYFSYKDNPKDISLQYALFTSPNVVVWDGSRG--YQNLFDAL 211
P++ + +P N Y +F+YKDNP ISL Y LF PN D + Y N+ A
Sbjct: 379 PLLSFHAQIKSPFLINAYPFFAYKDNPNQISLNYVLF-QPNQGATDPNTNLHYDNMLYAQ 555
Query: 212 LDSLHAAIDNTGIGFVKVVVSESGWPSDGG---FATTYDNARVYLDNLIRHV--KGGTPM 266
+D+++AAI G V+V +SE+GWPS G T NA +Y NL++ + K GTP
Sbjct: 556 IDAVYAAIKALGHTDVEVRISETGWPSKGDPDEVGATPQNAEIYNSNLLKRIEQKQGTPA 735
Query: 267 R-SGPIETYIFGLFDENQK-NPELEKHFGVFYPNKQKKYPFGFQG 309
S PI+ ++F LF+EN K P E+++G++YP+ Y G +G
Sbjct: 736 NPSVPIDIFVFALFNENLKPGPVSERNYGLYYPDGTPVYNIGLEG 870
>TC225887 similar to UP|Q9M4A9 (Q9M4A9) Beta-1,3 glucanase precursor ,
partial (95%)
Length = 1692
Score = 176 bits (446), Expect = 1e-44
Identities = 116/315 (36%), Positives = 175/315 (54%), Gaps = 10/315 (3%)
Frame = +3
Query: 2 MGNNLPSQREAIDLCKSNN-IKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
+G+NLP + K+ I R+++YD N L A SGI + + PN D+ + T
Sbjct: 165 LGDNLPPPAAVANFLKTKTTIDRVKIYDVNPDILRAFAGSGISVTVTAPNGDIAAL-TKI 341
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
D A QWV ++ F+P KI YI VG+EV G ++ V PA++ ++ A+ A+ + D
Sbjct: 342 DSARQWVATHIKPFHPQTKINYILVGSEVLHWGDTNMIRGLV-PAMRTLHSALLAEGITD 518
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSY-LDPIIGYLVYANAPLFANIYSYFSYKDN 179
IKV+TA + ++ +S PPS G FR + L P++ +L PL N Y YF Y N
Sbjct: 519 -IKVTTAHSLAIMRSSIPPSMGRFRPGYAKHVLGPMLKFLRETRTPLMVNPYPYFGY--N 689
Query: 180 PKDISLQYALFTSPNVVVWDG--SRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWP 237
K+++ L PN ++D R Y N FDAL+D++H+A++ G G V + V E+GWP
Sbjct: 690 GKNVNF---LLFRPNRGLYDRYTKRSYTNQFDALMDAVHSAMNALGYGDVDIAVGETGWP 860
Query: 238 S--DGGFATTYDNARVYLDNLIRHV---KGGTPMRSGPIETYIFGLFDENQK-NPELEKH 291
S DG A + NA+ + L++H+ KG M + ETYIF LF+ENQK P E++
Sbjct: 861 SVCDGWDACSVANAQSFNRELVKHLATGKGTPLMPNRSFETYIFALFNENQKPGPIAERN 1040
Query: 292 FGVFYPNKQKKYPFG 306
+G+F P+ Y G
Sbjct: 1041WGLFQPDFTPVYDSG 1085
>TC218883 weakly similar to UP|Q9MAQ2 (Q9MAQ2) CDS, partial (54%)
Length = 875
Score = 151 bits (381), Expect = 4e-37
Identities = 85/226 (37%), Positives = 132/226 (57%), Gaps = 2/226 (0%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ NNLPSQ +A+ L KS +++LYD + L+A N+G+ELM+G+ N L +
Sbjct: 202 IANNLPSQDDAVALVKSIGATKVKLYDADPRVLKAFANTGVELMVGLGNEYLSRMKDPKQ 381
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A W++ N+ + P+ KI I VGNEV +S + +LPA+Q+++ A+ L Q
Sbjct: 382 -AQAWIKANLQPYLPATKITSIFVGNEVLTFNDTSLTSN-LLPAMQSVHAALINLGLDKQ 555
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPK 181
I V+T + ++ TSYPPS G+FR D+ L PI+ + +P N Y YF+YK NPK
Sbjct: 556 ITVTTTHSLAVLQTSYPPSAGAFRPDLAPCLAPILSFQAKTGSPFLINAYPYFAYKANPK 735
Query: 182 DISLQYALFTSPNVVVWDGSRG--YQNLFDALLDSLHAAIDNTGIG 225
+ L+Y LF PN + D S Y N+ A +D++++A+D+ G G
Sbjct: 736 QVPLEYVLF-QPNEGMVDPSSNLHYDNMLFAQIDAVYSALDSLGYG 870
>BG352819 homologue to SP|Q03773|E13A Glucan endo-1 3-beta-glucosidase
precursor (EC 3.2.1.39) ((1->3)-beta- glucan
endohydrolase) ((1->3), partial (30%)
Length = 460
Score = 130 bits (327), Expect = 8e-31
Identities = 65/105 (61%), Positives = 81/105 (76%), Gaps = 1/105 (0%)
Frame = +2
Query: 141 KGSFRSDVR-SYLDPIIGYLVYANAPLFANIYSYFSYKDNPKDISLQYALFTSPNVVVWD 199
K SF+SD R +YLD +I +LV NAPL N+YSYF+Y NPKDISL YALF SP+VVV D
Sbjct: 146 KCSFKSDYRGAYLDGVIRFLVNNNAPLMVNVYSYFAYTANPKDISLDYALFRSPSVVVQD 325
Query: 200 GSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGGFAT 244
GS GY+NLFDA +D+++AA++ G + +VVSESGWPS GG AT
Sbjct: 326 GSLGYRNLFDASVDAVYAALEKAXGGSLNIVVSESGWPSSGGTAT 460
>TC217312 similar to GB|AAO63352.1|28950857|BT005288 At2g26600 {Arabidopsis
thaliana;} , partial (51%)
Length = 569
Score = 129 bits (323), Expect = 2e-30
Identities = 63/179 (35%), Positives = 106/179 (59%)
Frame = +2
Query: 21 IKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIAIQWVQKNVLNFYPSVKI 80
I+ +R+YD + + L+A +G+E+++G+PN LQ++++N D A+ WV++NV +F P +I
Sbjct: 11 IRNVRIYDADHSVLKAFSGTGLEIVVGLPNGQLQDMSSNPDHALNWVKENVQSFLPDTRI 190
Query: 81 KYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQIKVSTAIDMTMIGTSYPPS 140
+ IAVGNEV GG +L A++NIY A +L +++STA + SYPPS
Sbjct: 191 RGIAVGNEVLG-GGDYSLWGVLLGAVKNIYNATVKLHLDQLVQISTANSFAVFSQSYPPS 367
Query: 141 KGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPKDISLQYALFTSPNVVVWD 199
G F +V ++ P++ + +P N Y + Y +P+ I + YALF P ++D
Sbjct: 368 SGKFDDNVNQFMKPLLEFFQQIGSPFCVNAYPFLVYASDPEHIDINYALF-EPTKGIYD 541
>TC209407 UP|E13A_SOYBN (Q03773) Glucan endo-1,3-beta-glucosidase precursor
((1->3)-beta-glucan endohydrolase)
((1->3)-beta-glucanase) (Beta-1,3-endoglucanase) ,
partial (38%)
Length = 759
Score = 123 bits (309), Expect = 1e-28
Identities = 59/125 (47%), Positives = 89/125 (71%)
Frame = +2
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+GNNLP+ +E + L NI+RMR+Y P+ LEALR S IEL+L +PN +L+N+A++ D
Sbjct: 392 LGNNLPTPQEVVALYNQANIRRMRIYGPSPEVLEALRGSNIELLLDIPNDNLRNLASSQD 571
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A +WVQ N+ N+ +V+ +Y++VGNEV P FA+F++PA++NI +AI L +Q
Sbjct: 572 NANKWVQDNIKNYANNVRFRYVSVGNEVKP---EHSFAQFLVPALENIQRAISNAGLGNQ 742
Query: 122 IKVST 126
+KVST
Sbjct: 743 VKVST 757
>TC224124 similar to UP|Q8L9D9 (Q8L9D9) Beta-1,3-glucanase-like protein,
partial (38%)
Length = 532
Score = 112 bits (281), Expect = 2e-25
Identities = 67/167 (40%), Positives = 95/167 (56%), Gaps = 7/167 (4%)
Frame = +3
Query: 108 NIYQAIRAKNLHDQIKVSTAIDMTMIGTSYPPSKGSFRSD-VRSYLDPIIGYLVYANAPL 166
NI +A+ NL IKVS+ I ++ + SYP S GSFR + V P++ +L + L
Sbjct: 3 NIQKALTKHNLDKDIKVSSPIALSALANSYPSSAGSFRPELVEPVFKPMLDFLRETGSYL 182
Query: 167 FANIYSYFSYKDNPKDISLQYALFT-SPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIG 225
N+Y +F+Y+ N ISL YALF +P VV Y NLFDA +D++ +A+
Sbjct: 183 MVNVYPFFAYESNADVISLDYALFRDNPGVVDPGNGLRYYNLFDAQIDAVFSALSALKYD 362
Query: 226 FVKVVVSESGWPSDGG---FATTYDNARVYLDNLIRHV--KGGTPMR 267
VK+VV+E+GWPS G + DNA Y NL+R + GGTP+R
Sbjct: 363 DVKIVVTETGWPSKGDSNEVGASVDNAAAYNGNLVRKILTAGGTPLR 503
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,471,650
Number of Sequences: 63676
Number of extensions: 152052
Number of successful extensions: 774
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 699
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 699
length of query: 327
length of database: 12,639,632
effective HSP length: 98
effective length of query: 229
effective length of database: 6,399,384
effective search space: 1465458936
effective search space used: 1465458936
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144539.10


