

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144517.6 + phase: 0
(366 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
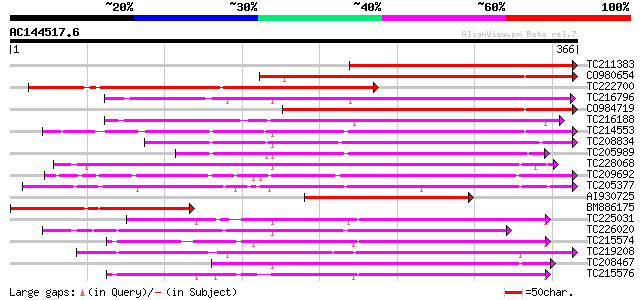
Score E
Sequences producing significant alignments: (bits) Value
TC211383 weakly similar to UP|Q8L812 (Q8L812) Fruit ripening-rel... 254 6e-68
CO980654 207 8e-54
TC222700 similar to GB|AAS76251.1|45752706|BT012156 At1g55290 {A... 203 9e-53
TC216796 similar to GB|AAQ65160.1|34365697|BT010537 At4g10500 {A... 203 1e-52
CO984719 201 4e-52
TC216188 similar to UP|Q84L58 (Q84L58) 1-aminocyclopropane-1-car... 174 7e-44
TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 174 7e-44
TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 170 8e-43
TC205989 similar to UP|Q9FLV0 (Q9FLV0) Flavanone 3-hydroxylase-l... 162 3e-40
TC228068 anthocyanidin synthase [Glycine max] 160 6e-40
TC209692 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1... 158 3e-39
TC205377 154 5e-38
AI930725 weakly similar to SP|P10967|ACC3_ 1-aminocyclopropane-1... 151 5e-37
BM886175 homologue to GP|28949891|emb 1-aminocyclopropane-1-carb... 151 5e-37
TC225031 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-c... 150 9e-37
TC226020 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, ... 150 9e-37
TC215574 similar to UP|O04076 (O04076) ACC-oxidase, partial (98%) 147 6e-36
TC219208 weakly similar to UP|Q948K9 (Q948K9) CmE8 protein, part... 146 2e-35
TC208467 similar to UP|Q8LP22 (Q8LP22) Flavonol synthase, partia... 145 4e-35
TC215576 similar to UP|O04076 (O04076) ACC-oxidase, partial (97%) 142 2e-34
>TC211383 weakly similar to UP|Q8L812 (Q8L812) Fruit ripening-related ACC
oxidase (Fragment), partial (21%)
Length = 555
Score = 254 bits (648), Expect = 6e-68
Identities = 121/147 (82%), Positives = 136/147 (92%)
Frame = +3
Query: 220 MLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVL 279
M+AAN+YPPCPQP+LAMG+PPHSDHGLLNLL+QNGVSGLQVLHNGKWINV STSNC LV
Sbjct: 3 MIAANMYPPCPQPELAMGIPPHSDHGLLNLLLQNGVSGLQVLHNGKWINVGSTSNCQLVF 182
Query: 280 VSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYV 339
VSDHLE++SNGKYKSV+HRA VSN ATRMSLA VIAPSLDTVVEPA E LDN+ NPAAYV
Sbjct: 183 VSDHLEVVSNGKYKSVLHRAVVSNKATRMSLAVVIAPSLDTVVEPAKEFLDNQRNPAAYV 362
Query: 340 GMKHIDYMKLQRNNQLYGKSVLNKVKI 366
GMKH DYM+LQ++N+L GKSVL++VKI
Sbjct: 363 GMKHRDYMQLQKSNRLNGKSVLDRVKI 443
>CO980654
Length = 844
Score = 207 bits (526), Expect = 8e-54
Identities = 110/231 (47%), Positives = 146/231 (62%), Gaps = 26/231 (11%)
Frame = -1
Query: 162 VHPKFHSPDKPSGFR--------------------------ETSAEYSRKTWKLGRELLK 195
VHP F+ P KP GF ET EY K+ ++ ELLK
Sbjct: 838 VHPHFNVPSKPHGFSRDWWVQTKC*CVCEVSFVLAPKNHACETVDEYITKSREVVGELLK 659
Query: 196 GISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGV 255
GIS SLGLE NYI K +N++ G Q L N YPPCP+P+L MG+P H+DHGLL LL++N +
Sbjct: 658 GISLSLGLEENYIHKRLNVELGSQFLILNFYPPCPKPELVMGLPAHTDHGLLTLLMENEL 479
Query: 256 SGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIA 315
GLQ+ H G+WI V + N FL+ DHLEI++NGKYKSV+HRA V+ ATR+S+AT
Sbjct: 478 GGLQIQHKGRWIPVHALPNSFLINTGDHLEILTNGKYKSVLHRAVVNTKATRISVATAHG 299
Query: 316 PSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
LDT V PA EL+ +E NPAAY +K+ DY+ Q++N+L +S L+ ++I
Sbjct: 298 APLDTSVGPAPELVGDE-NPAAYRAIKYRDYIHFQQSNELDRRSCLDHIRI 149
>TC222700 similar to GB|AAS76251.1|45752706|BT012156 At1g55290 {Arabidopsis
thaliana;} , partial (8%)
Length = 677
Score = 203 bits (517), Expect = 9e-53
Identities = 106/226 (46%), Positives = 144/226 (62%)
Frame = +2
Query: 13 SNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLIN 72
S + +SVK L ES +S+PS+Y NP D + + + D IP ID+S L +
Sbjct: 17 SREKYSKMSSVKELVESKCLSSVPSNYICLENPEDS---ILNYE--TDNIPTIDFSQLTS 181
Query: 73 GNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKE 132
N R+K I +G AC +WGFF+L NH VS+ L ++++ FF+L E++K +A +
Sbjct: 182 SNPSVRSKAIKQLGDACRDWGFFMLINHGVSEILRDEVIRTSQGFFDLTEKEKMEHAGRN 361
Query: 133 VTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPSGFRETSAEYSRKTWKLGRE 192
+ D I+YGTSFNV+ DK LFWRD++K VHP F++P KP GF +T EY K +L E
Sbjct: 362 LFDP-IRYGTSFNVTVDKTLFWRDYLKCHVHPHFNAPSKPPGFSQTLEEYITKGRELVEE 538
Query: 193 LLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGM 238
LL+GIS SLGLE N+I K MNLD G Q+L N YPPCP P+L MG+
Sbjct: 539 LLEGISLSLGLEENFIHKRMNLDLGSQLLVINCYPPCPNPELVMGL 676
>TC216796 similar to GB|AAQ65160.1|34365697|BT010537 At4g10500 {Arabidopsis
thaliana;} , partial (72%)
Length = 1454
Score = 203 bits (516), Expect = 1e-52
Identities = 119/313 (38%), Positives = 178/313 (56%), Gaps = 9/313 (2%)
Frame = +1
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK 121
IP+ID L N R+ I I +AC+ +GFF +TNH V + ++EK++ FF L
Sbjct: 214 IPLIDLQDLHGPN---RSHIIQQIDQACQNYGFFQVTNHGVPEGVIEKIMKVTREFFGLP 384
Query: 122 EEDKQVYADKEVTDDSIK---YGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDKPSG 174
E +K K + D K TSFNV+ +K WRDF+++ HP P P
Sbjct: 385 ESEKL----KSYSTDPFKASRLSTSFNVNSEKVSSWRDFLRLHCHPIEDYIKEWPSNPPS 552
Query: 175 FRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGL--QMLAANLYPPCPQP 232
RE AEY RK + +L++ ISESLGLE +YI++ + G Q LA N YP CP+P
Sbjct: 553 LREDVAEYCRKMRGVSLKLVEAISESLGLERDYINRVVGGKKGQEQQHLAMNYYPACPEP 732
Query: 233 DLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKY 292
+L G+P H+D ++ +L+Q+ V GLQVL +GKW+ V+ N F+V V D ++++SN KY
Sbjct: 733 ELTYGLPGHTDPTVITILLQDEVPGLQVLKDGKWVAVNPIPNTFVVNVGDQIQVISNDKY 912
Query: 293 KSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRN 352
KSV+HRA V+ R+S+ T PS D ++ PA +L+ + +P Y + +Y + N
Sbjct: 913 KSVLHRAVVNCNKDRISIPTFYFPSNDAIIGPAPQLIHHHHHPPQYNNFTYNEYYQNFWN 1092
Query: 353 NQLYGKSVLNKVK 365
L ++ L+ K
Sbjct: 1093RGLSKETCLDIFK 1131
>CO984719
Length = 839
Score = 201 bits (511), Expect = 4e-52
Identities = 101/190 (53%), Positives = 136/190 (71%)
Frame = -1
Query: 177 ETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAM 236
ET EY K+ ++ ELLKGIS SLGLE NYI K +N++ G Q L N YPPCP+P+L M
Sbjct: 668 ETVDEYITKSREVVGELLKGISLSLGLEENYIHKRLNVELGSQFLILNFYPPCPKPELVM 489
Query: 237 GMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVV 296
G+P H+DHGLL LL++N + GLQ+ H G+WI V + N FL+ DHLEI++NGKYKSV+
Sbjct: 488 GLPAHTDHGLLTLLMENELGGLQIQHKGRWIPVHALPNSFLINTGDHLEILTNGKYKSVL 309
Query: 297 HRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLY 356
HRA V+ ATR+S+AT LDT V PA EL+ +E NPAAY +K+ DY+ Q++N+L
Sbjct: 308 HRAVVNTKATRISVATAHGAPLDTSVGPAPELVGDE-NPAAYRAIKYRDYIHFQQSNELD 132
Query: 357 GKSVLNKVKI 366
+S L+ ++I
Sbjct: 131 RRSCLDHIRI 102
>TC216188 similar to UP|Q84L58 (Q84L58) 1-aminocyclopropane-1-carboxylic acid
oxidase, complete
Length = 1362
Score = 174 bits (440), Expect = 7e-44
Identities = 105/304 (34%), Positives = 161/304 (52%), Gaps = 7/304 (2%)
Frame = +3
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNL- 120
+PVID+S L N ++RTKT+ I CEEWGFF L NH + + L+E++ F+ L
Sbjct: 147 VPVIDFSKL---NGEERTKTMAQIANGCEEWGFFQLINHGIPEELLERVKKVASEFYKLE 317
Query: 121 KEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPSGFRETSA 180
+EE+ + ++ DS++ +S D W D I ++ ++ P+K GFRET A
Sbjct: 318 REENFKNSTSVKLLSDSVEKKSSEMEHVD----WEDVITLLDDNEW--PEKTPGFRETMA 479
Query: 181 EYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQML---AANLYPPCPQPDLAMG 237
EY + KL +L++ + E+LGL YI K +N G + YPPCP P+L G
Sbjct: 480 EYRAELKKLAEKLMEVMDENLGLTKGYIKKALNGGDGENAFFGTKVSHYPPCPHPELVKG 659
Query: 238 MPPHSDHGLLNLLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVV 296
+ H+D G + LL Q+ V GLQ+L G+WI+V N ++ D +E++SNG+YKS
Sbjct: 660 LRAHTDAGGVILLFQDDKVGGLQMLKEGQWIDVQPLPNAIVINTGDQIEVLSNGRYKSCW 839
Query: 297 HRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHI--DYMKLQRNNQ 354
HR + R S+A+ PS + PA +L++ E K + DYM + +
Sbjct: 840 HRVLATPDGNRRSIASFYNPSFKATICPAPQLVEKEDQQVDETYPKFVFGDYMSVYAEQK 1019
Query: 355 LYGK 358
K
Sbjct: 1020FLPK 1031
>TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (52%)
Length = 1808
Score = 174 bits (440), Expect = 7e-44
Identities = 120/354 (33%), Positives = 181/354 (50%), Gaps = 9/354 (2%)
Frame = +1
Query: 22 SVKTLSESPDFNSIPSSYTYTT-NPHD-ENEIVADQDEVNDPIPVIDYSLLINGNHDQRT 79
SV L++ P +P Y + +PH N I Q +P+ID L++ + +
Sbjct: 61 SVLELAKQPIIE-VPERYVHANQDPHILSNTISLPQ------VPIIDLHQLLSEDPSELE 219
Query: 80 KTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIK 139
K H AC+EWGFF L NH V ++E M V FFNL E+KQ + + +D
Sbjct: 220 KLDH----ACKEWGFFQLINHGVDPPVVENMKIGVQEFFNLPMEEKQKFW--QTPEDMQG 381
Query: 140 YGTSFNVSGDKNLFWRDFIKIIVHPKFHS------PDKPSGFRETSAEYSRKTWKLGREL 193
+G F VS ++ L W D P HS P P FRE Y + K+ +
Sbjct: 382 FGQLFVVSEEQKLEWADMFYAHTFP-LHSRNPHLIPKIPQPFRENLENYCLELRKMCITI 558
Query: 194 LKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ- 252
+ + ++L ++ N + + S Q + N YPPCPQP+ +G+ PHSD G L +L+Q
Sbjct: 559 IGLMKKALKIKTNELSELFEDPS--QGIRMNYYPPCPQPERVIGINPHSDSGALTILLQV 732
Query: 253 NGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLAT 312
N V GLQ+ +GKWI V SN F++ V D LEI++NG Y+S+ HR V++ R+S+A
Sbjct: 733 NEVEGLQIRKDGKWIPVKPLSNAFVINVGDMLEILTNGIYRSIEHRGIVNSEKERISIAM 912
Query: 313 VIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
P + V+ PA L+ E PA + + DY+ +L GKS ++ ++I
Sbjct: 913 FHRPQMSRVIGPAPSLVTPE-RPALFKRIGVADYLNGFLKRELKGKSYMDVIRI 1071
>TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (54%)
Length = 1070
Score = 170 bits (431), Expect = 8e-43
Identities = 107/287 (37%), Positives = 160/287 (55%), Gaps = 8/287 (2%)
Frame = +2
Query: 88 ACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVS 147
AC+EWGFF L NH V+ SL+EK+ + FFNL +K+ + + +G +F VS
Sbjct: 44 ACKEWGFFQLVNHGVNSSLVEKVRLETQDFFNLPMSEKKKFW--QTPQHMEGFGQAFVVS 217
Query: 148 GDKNLFWRDFIKIIVHPKFHS------PDKPSGFRETSAEYSRKTWKLGRELLKGISESL 201
D+ L W D + PK HS P P FR+T YSR+ L ++ + ++L
Sbjct: 218 EDQKLDWADLYYMTTLPK-HSRMPHLFPQLPLPFRDTLEAYSREIKDLAIVIIGLMGKAL 394
Query: 202 GLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDH-GLLNLLIQNGVSGLQV 260
++ I + + G+Q++ N YPPCP+P+ +G+ PHSD GL LL N V GLQ+
Sbjct: 395 KIQEREIRELF--EDGIQLMRMNYYPPCPEPEKVIGLTPHSDGIGLAILLQLNEVEGLQI 568
Query: 261 LHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDT 320
+G W+ V N F+V V D LEI++NG Y+S+ HRA V+ R+S AT +PS D
Sbjct: 569 RKDGLWVPVKPLINAFIVNVGDILEIITNGIYRSIEHRATVNGEKERLSFATFYSPSSDG 748
Query: 321 VVEPASELLDNESNPA-AYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
VV PA L+ ++ P +G+K DY K + +L GK+ + ++I
Sbjct: 749 VVGPAPSLITEQTPPRFKSIGVK--DYFKGLFSRKLDGKAYIEVMRI 883
>TC205989 similar to UP|Q9FLV0 (Q9FLV0) Flavanone 3-hydroxylase-like protein,
partial (74%)
Length = 1192
Score = 162 bits (409), Expect = 3e-40
Identities = 94/247 (38%), Positives = 146/247 (59%), Gaps = 6/247 (2%)
Frame = +2
Query: 108 EKMVDQVFAFFNLKEEDK-QVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHP-- 164
++M + FF L E+K ++Y+ E T +++ TSFNV + WRD++++ +P
Sbjct: 5 KEMEEVAHGFFKLPVEEKLKLYS--EDTSKTMRLSTSFNVKKETVRNWRDYLRLHCYPLE 178
Query: 165 KFHS--PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLA 222
K+ P P F+ET EY +LG + + ISESLGLE +YI + Q +A
Sbjct: 179 KYAPEWPSNPPSFKETVTEYCTIIRELGLRIQEYISESLGLEKDYIKNVLGEQG--QHMA 352
Query: 223 ANLYPPCPQPDLAMGMPPHSDHGLLNLLIQN-GVSGLQVLHNGKWINVSSTSNCFLVLVS 281
N YPPCP+P+L G+P H+D L +L+Q+ V+GLQVL +GKW+ VS N F++ +
Sbjct: 353 VNYYPPCPEPELTYGLPGHTDPNALTILLQDLQVAGLQVLKDGKWLAVSPQPNAFVINIG 532
Query: 282 DHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGM 341
D L+ +SNG YKSV HRA V+ R+S+A+ + P+ + ++ PA L ++ S A Y G
Sbjct: 533 DQLQALSNGLYKSVWHRAVVNVEKPRLSVASFLCPNDEALISPAKPLTEHGSE-AVYRGF 709
Query: 342 KHIDYMK 348
+ +Y K
Sbjct: 710 TYAEYYK 730
>TC228068 anthocyanidin synthase [Glycine max]
Length = 1231
Score = 160 bits (406), Expect = 6e-40
Identities = 103/340 (30%), Positives = 169/340 (49%), Gaps = 14/340 (4%)
Frame = +3
Query: 29 SPDFNSIPSSYTYTTNPHDE----NEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHD 84
S IP Y P +E + ++ + +P ID + + + R K
Sbjct: 39 SSGIKCIPKEYV---RPQEELKSIGNVFEEEKKEGLQVPTIDLREIDSEDEVVRGKCREK 209
Query: 85 IGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSF 144
+ KA EEWG L NH + L+E++ FF L E+K+ YA+ + YG+
Sbjct: 210 LKKAAEEWGVMHLVNHGIPDELIERVKKAGETFFGLAVEEKEKYANDLESGKIQGYGSKL 389
Query: 145 NVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLKGISE 199
+ L W D+ + P+ P KP+ + E ++EY+++ L ++L+ +S
Sbjct: 390 ANNASGQLEWEDYFFHLAFPEDKRDLSFWPKKPADYIEVTSEYAKRLRGLATKILEALSI 569
Query: 200 SLGLEVNYIDKTMN-LDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGL 258
LGLE ++K + ++ L L N YP CPQP+LA+G+ H+D L L+ N V GL
Sbjct: 570 GLGLEGRRLEKEVGGMEELLLQLKINYYPICPQPELALGVEAHTDVSSLTFLLHNMVPGL 749
Query: 259 QVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSL 318
Q+ + G+W+ N L+ + D +EI+SNGKYKS++HR V+ R+S A P
Sbjct: 750 QLFYQGQWVTAKCVPNSILMHIGDTIEILSNGKYKSILHRGLVNKEKVRISWAVFCEPPK 929
Query: 319 DTVV-EPASELLDNESNPAAY---VGMKHIDYMKLQRNNQ 354
+ ++ +P EL+ E+ PA + +HI + KL R +Q
Sbjct: 930 EKIILQPLPELV-TETEPARFPPRTFAQHIHH-KLFRKDQ 1043
>TC209692 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (36%)
Length = 1229
Score = 158 bits (400), Expect = 3e-39
Identities = 117/352 (33%), Positives = 180/352 (50%), Gaps = 8/352 (2%)
Frame = +2
Query: 23 VKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTI 82
++ L + P S+P Y N +E ++A + + +P I+ LI+G + +
Sbjct: 32 IQELIKKP-LTSVPQRYIQLHN--NEPSLLAGET-FSHALPTINLKKLIHGEDIELE--L 193
Query: 83 HDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGT 142
+ AC +WGFF L H +S +M+ + D+V FF L E+K Y K D YGT
Sbjct: 194 EKLTSACRDWGFFQLVEHGISSVVMKTLEDEVEGFFMLPMEEKMKY--KVRPGDVEGYGT 367
Query: 143 SFNVSGDKNLFWRD--FIKI----IVHPKFHSPDKPSGFRETSAEYSRKTWKLGRELLKG 196
S D+ L W D F+KI I +P P+ PS R Y + L L+
Sbjct: 368 VIG-SEDQKLDWGDRLFMKINXRSIRNPHLF-PELPSSLRNILELYIEELQNLAMILMGL 541
Query: 197 ISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ-NGV 255
+ ++L +E ++ + G+Q + YPPCPQP+L MG+ HSD + +L Q NGV
Sbjct: 542 LGKTLKIEKRELEV---FEDGIQNMRMTYYPPCPQPELVMGLSAHSDATGITILNQMNGV 712
Query: 256 SGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIA 315
+GLQ+ +G WI V+ S +V + D +EIMSNG YKSV HRA V++ R+S+A
Sbjct: 713 NGLQIKKDGVWIPVNVISEALVVNIGDIIEIMSNGAYKSVEHRATVNSEKERISVAMFFL 892
Query: 316 PSLDTVVEPASELLDNESNPAAYVGMKHIDYMK-LQRNNQLYGKSVLNKVKI 366
P + + PA L N +P + + +Y+K +N+L GKS L ++I
Sbjct: 893 PKFQSEIGPAVS-LTNPEHPPLFKRIVVEEYIKDYFTHNKLNGKSYLEHMRI 1045
>TC205377
Length = 1345
Score = 154 bits (390), Expect = 5e-38
Identities = 114/382 (29%), Positives = 185/382 (47%), Gaps = 24/382 (6%)
Frame = +3
Query: 9 VNQKSNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYS 68
+N+++ ++ +T K L++SP + +++ +P ++ Q E IP+ID S
Sbjct: 21 LNKRTAQNLSKYTCCKLLTQSPAMGEVDAAFIQ--DPPHRPKLSTIQAE---GIPIIDLS 185
Query: 69 LLINGNHDQRTKT---IHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDK 125
+ N + + +IG AC EWGFF +TNH V +L + + FF E+K
Sbjct: 186 PITNHRVSDPSAIEGLVKEIGSACNEWGFFQVTNHGVPLTLRQNIEKASKLFFAQSAEEK 365
Query: 126 QVYADKEVTDDSIKYGTSF--NVSGDKNLFWRDFIKIIVHPKF----------------- 166
+ + E + Y T NV K +F DF+ P F
Sbjct: 366 RKVSRNESSPAGY-YDTEHTKNVRDWKEVF--DFLA--KEPTFIPVTSDEHDDRVNQWTN 530
Query: 167 HSPDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLY 226
SP+ P FR + EY ++ KL ++L+ I+ SLGLE +++ D + N Y
Sbjct: 531 QSPEYPLNFRVVTQEYIQEMEKLSFKILELIALSLGLEAKRVEEFFIKDQ-TSFIRLNHY 707
Query: 227 PPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNG--KWINVSSTSNCFLVLVSDHL 284
PPCP PDLA+G+ H D G L +L Q+ V GL+V +WI V T + +++ + D +
Sbjct: 708 PPCPYPDLALGVGRHKDPGALTILAQDEVGGLEVRRKADQEWIRVKPTPDAYIINIGDTV 887
Query: 285 EIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHI 344
++ SN Y+SV HR V++ R S+ P+ DT V+P EL+ NE NP+ Y K
Sbjct: 888 QVWSNDAYESVDHRVVVNSEKERFSIPFFFFPAHDTEVKPLEELI-NEQNPSKYRPYKWG 1064
Query: 345 DYMKLQRNNQLYGKSVLNKVKI 366
++ + R N + K ++I
Sbjct: 1065KFL-VHRGNSNFKKQNEENIQI 1127
>AI930725 weakly similar to SP|P10967|ACC3_ 1-aminocyclopropane-1-carboxylate
oxidase homolog (Protein E8). [Tomato] {Lycopersicon
esculentum}, partial (20%)
Length = 342
Score = 151 bits (381), Expect = 5e-37
Identities = 70/109 (64%), Positives = 86/109 (78%)
Frame = +1
Query: 191 RELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLL 250
R+LL+GISESLGLE N I ++ + DSG Q+ NLYPPCPQP LA+G+P HSD GLL LL
Sbjct: 16 RKLLEGISESLGLESNSIIESTDFDSGHQLFVVNLYPPCPQPHLALGLPSHSDVGLLTLL 195
Query: 251 IQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRA 299
QNG+ GLQV HNGKW+NV+ NC +VL+SD LE++SNGKY V+HRA
Sbjct: 196 TQNGIGGLQVKHNGKWVNVNPLPNCLIVLLSDQLEVVSNGKYARVMHRA 342
>BM886175 homologue to GP|28949891|emb 1-aminocyclopropane-1-carboxylic acid
oxidase {Cicer arietinum}, partial (3%)
Length = 422
Score = 151 bits (381), Expect = 5e-37
Identities = 79/120 (65%), Positives = 91/120 (75%), Gaps = 1/120 (0%)
Frame = +2
Query: 1 MASTLPPQVNQ-KSNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVN 59
MAST Q +SNN F SVK L+ESP+ S+P SYTYTTN DE IVAD DE +
Sbjct: 71 MASTASEVSQQVQSNNNRPIFKSVKALTESPELTSLPPSYTYTTNSDDE--IVADPDE-D 241
Query: 60 DPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFN 119
DPIP+IDYSLL+ G DQR TIHD+GKACEEWGFF+L NH VSK++MEKMVD+VFAFFN
Sbjct: 242 DPIPIIDYSLLVTGTPDQRAMTIHDLGKACEEWGFFMLINHFVSKTIMEKMVDEVFAFFN 421
>TC225031 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-carboxylic
acid oxidase, complete
Length = 1244
Score = 150 bits (379), Expect = 9e-37
Identities = 95/284 (33%), Positives = 150/284 (52%), Gaps = 10/284 (3%)
Frame = +1
Query: 76 DQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFN--LKEEDKQVYADKEV 133
++R T+ I ACE WGFF L NH + +++ + + ++E K++ A K +
Sbjct: 154 EERNDTMEKIKDACENWGFFELVNHGIPHDILDTVERLTKEHYRKCMEERFKELVASKGL 333
Query: 134 TDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS---PDKPSGFRETSAEYSRKTWKLG 190
D+++ K++ W + P+ + PD +R+ +++ + KL
Sbjct: 334 --DAVQTEV-------KDMDWESTFHLRHLPESNISEIPDLIDEYRKVMKDFALRLEKLA 486
Query: 191 RELLKGISESLGLEVNYIDKTMNLDSG--LQMLAANLYPPCPQPDLAMGMPPHSDHGLLN 248
+LL + E+LGLE Y+ K G AN YPPCP P+L G+ PH+D G +
Sbjct: 487 EQLLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVAN-YPPCPNPELVKGLRPHTDAGGII 663
Query: 249 LLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATR 307
LL Q+ VSGLQ+L +G+W++V + +V + D LE+++NGKYKSV HR TR
Sbjct: 664 LLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNIGDQLEVITNGKYKSVEHRVIAQTDGTR 843
Query: 308 MSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHI--DYMKL 349
MS+A+ P D V+ PA ELL+ E+ + K + DYMKL
Sbjct: 844 MSIASFYNPGSDAVIYPAPELLEKEAEEKNQLYPKFVFEDYMKL 975
>TC226020 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, partial
(93%)
Length = 1441
Score = 150 bits (379), Expect = 9e-37
Identities = 100/311 (32%), Positives = 161/311 (51%), Gaps = 8/311 (2%)
Frame = +3
Query: 22 SVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKT 81
+ KTL+ ++ SS+ +E VA +E +D IPVI + I+ +R +
Sbjct: 72 TAKTLTYLAQEKTLESSFV---RDEEERPKVA-YNEFSDEIPVISLAG-IDEVDGRRREI 236
Query: 82 IHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYG 141
I +ACE WG F + +H V + L+ +M FF L ++K + +
Sbjct: 237 CEKIVEACENWGIFQVVDHGVDQQLVAEMTRLAKEFFALPPDEKLRFDMSGAKKGG--FI 410
Query: 142 TSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLKG 196
S ++ G+ WR+ + +PK PD P G+R + EYS K L +L++
Sbjct: 411 VSSHLQGESVQDWREIVTYFSYPKRERDYSRWPDTPEGWRSVTEEYSDKVMGLACKLMEV 590
Query: 197 ISESLGLEVNYIDKT-MNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGV 255
+SE++GLE + K +++D Q + N YP CPQPDL +G+ H+D G + LL+Q+ V
Sbjct: 591 LSEAMGLEKEGLSKACVDMD---QKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQV 761
Query: 256 SGLQVLH-NGK-WINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATV 313
GLQ NGK WI V F+V + DH +SNG++K+ H+A V++ +R+S+AT
Sbjct: 762 GGLQATRDNGKTWITVQPVEAAFVVNLGDHAHYLSNGRFKNADHQAVVNSNHSRLSIATF 941
Query: 314 IAPSLDTVVEP 324
P+ + V P
Sbjct: 942 QNPAPNATVYP 974
>TC215574 similar to UP|O04076 (O04076) ACC-oxidase, partial (98%)
Length = 1169
Score = 147 bits (372), Expect = 6e-36
Identities = 98/295 (33%), Positives = 153/295 (51%), Gaps = 8/295 (2%)
Frame = +3
Query: 63 PVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKE 122
PVI+ L N N ++R T++ I AC+ WGFF L NH + L++ + L +
Sbjct: 51 PVIN---LENLNGEERKATLNQIEDACQNWGFFELVNHGIPLELLDTVE-------RLTK 200
Query: 123 EDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRD--FIKIIVHPKFHS-PDKPSGFRETS 179
E + +K + G V K++ W F++ + PD +R+
Sbjct: 201 EHYRKCMEKRFKEAVSSKGLEAEV---KDMDWESTFFLRHLPTSNISEIPDLSQEYRDAM 371
Query: 180 AEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQM--LAANLYPPCPQPDLAMG 237
E+++K KL ELL + E+LGLE Y+ G AN YP CP+P+L G
Sbjct: 372 KEFAQKLEKLAEELLDLLCENLGLEKGYLKNAFYGSRGPNFGTKVAN-YPACPKPELVKG 548
Query: 238 MPPHSDHGLLNLLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVV 296
+ H+D G + LL+Q+ VSGLQ+L NG+W++V + +V + D +E+++NG+YKSV
Sbjct: 549 LRAHTDAGGIILLLQDDKVSGLQLLKNGQWVDVPPMRHSIVVNLGDQIEVITNGRYKSVE 728
Query: 297 HRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHI--DYMKL 349
HR TRMS+A+ P+ D ++ PA LL+ ++ V K + DYMKL
Sbjct: 729 HRVIAQTNGTRMSVASFYNPASDALIYPAPALLEQKAEDTEQVYPKFVFEDYMKL 893
>TC219208 weakly similar to UP|Q948K9 (Q948K9) CmE8 protein, partial (87%)
Length = 1242
Score = 146 bits (368), Expect = 2e-35
Identities = 103/330 (31%), Positives = 163/330 (49%), Gaps = 7/330 (2%)
Frame = +1
Query: 44 NPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVS 103
+P DE + + IPVID + + + R + I I +A E WGFF + NH +
Sbjct: 166 HPPDEFVMASKLGNTEYTIPVIDLAE-VGKDPSSRQEIIGRIREASERWGFFQVVNHGIP 342
Query: 104 KSLMEKMVDQVFAFFNLK-EEDKQVYADKEVTDDSIK---YGTSFNVSGDKNLFWRDFIK 159
+++E + D V F EE K++Y T D +K Y ++F++ L WRD
Sbjct: 343 VTVLEDLKDGVQRFHEQDIEEKKELY-----TRDHMKPLVYNSNFDLYSSPALNWRDSFM 507
Query: 160 IIVHPKFHSP-DKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGL 218
+ P P D P R+ EY KLG L + +SE+LGL +++ K + GL
Sbjct: 508 CYLAPNPPKPEDLPVVCRDILLEYGTYVMKLGIALFELLSEALGLHPDHL-KDLGCAEGL 684
Query: 219 QMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLV 278
L + YP CP+PDL +G HSD+ L +L+Q+ + GLQVL+ WI+++ +V
Sbjct: 685 ISLC-HYYPACPEPDLTLGTTKHSDNCFLTVLLQDHIGGLQVLYQNMWIDITPEPGALVV 861
Query: 279 LVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASEL--LDNESNPA 336
+ D L++++N ++KSV HR + R+S+A + L + +P + L E NP
Sbjct: 862 NIGDLLQLITNDRFKSVEHRVQANLIGPRISVACFFSEGLKSSPKPYGPIKELLTEDNPP 1041
Query: 337 AYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
Y +Y++ L G S L KI
Sbjct: 1042KYRETTVAEYVRYFEAKGLDGTSALQHFKI 1131
>TC208467 similar to UP|Q8LP22 (Q8LP22) Flavonol synthase, partial (68%)
Length = 933
Score = 145 bits (365), Expect = 4e-35
Identities = 84/228 (36%), Positives = 116/228 (50%), Gaps = 6/228 (2%)
Frame = +1
Query: 131 KEVTDDSIK-YGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSR 184
K DSI+ YGT + W D + IV P P P +RE + EY +
Sbjct: 13 KPAGSDSIEGYGTKLQKEVNGKKGWVDHLFHIVWPPSSINYSFWPQNPPSYREVNEEYCK 192
Query: 185 KTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDH 244
+ +L K +S LGLE N + + N D +L N YPPCP PDL +G+PPH+D
Sbjct: 193 HLRGVVDKLFKSMSVGLGLEENELKEGANEDDMHYLLKINYYPPCPCPDLVLGVPPHTDM 372
Query: 245 GLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNG 304
L +L+ N V GLQ +G W +V N ++ + D +EI+SNGKYK+V HR V+
Sbjct: 373 SYLTILVPNEVQGLQACRDGHWYDVKYVPNALVIHIGDQMEILSNGKYKAVFHRTTVNKD 552
Query: 305 ATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRN 352
TRMS I P + V P +L+ N+ NP Y K+ DY + N
Sbjct: 553 ETRMSWPVFIEPKKEQEVGPHPKLV-NQDNPPKYKTKKYKDYAYCKLN 693
>TC215576 similar to UP|O04076 (O04076) ACC-oxidase, partial (97%)
Length = 1151
Score = 142 bits (359), Expect = 2e-34
Identities = 98/296 (33%), Positives = 155/296 (52%), Gaps = 9/296 (3%)
Frame = +2
Query: 63 PVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFN--L 120
PVI+ L N N ++R T+ I AC+ WGFF L NH + L++ + + +
Sbjct: 2 PVIN---LDNLNGEERKATLDQIEDACQNWGFFELVNHGIPLELLDTVERLTKEHYRKCM 172
Query: 121 KEEDKQVYADK--EVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPSGFRET 178
+ K+ A K EV + + ++F + ++L + +I PD +R+
Sbjct: 173 ENRFKEAVASKALEVEVKDMDWESTFFL---RHLPTSNISEI--------PDLSQEYRDA 319
Query: 179 SAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQM--LAANLYPPCPQPDLAM 236
E+++K KL ELL + E+LGLE Y+ G AN YP CP+P+L
Sbjct: 320 MKEFAKKLEKLAEELLDLLCENLGLEKGYLKNAFYGSKGPNFGTKVAN-YPACPKPELVK 496
Query: 237 GMPPHSDHGLLNLLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSV 295
G+ H+D G + LL+Q+ VSGLQ+L + +W++V + +V + D +E+++NG+YKSV
Sbjct: 497 GLRAHTDAGGIILLLQDDKVSGLQLLKDDQWVDVPPMRHSIVVNLGDQIEVITNGRYKSV 676
Query: 296 VHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHI--DYMKL 349
HR TRMS+A+ P+ D V+ PA LL+ E+ V K + DYMKL
Sbjct: 677 EHRVVARTDGTRMSVASFYNPANDAVIYPAPALLEKEAQETEQVYPKFVFEDYMKL 844
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,954,038
Number of Sequences: 63676
Number of extensions: 228157
Number of successful extensions: 1255
Number of sequences better than 10.0: 211
Number of HSP's better than 10.0 without gapping: 1117
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1123
length of query: 366
length of database: 12,639,632
effective HSP length: 99
effective length of query: 267
effective length of database: 6,335,708
effective search space: 1691634036
effective search space used: 1691634036
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144517.6


