

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144516.12 - phase: 0
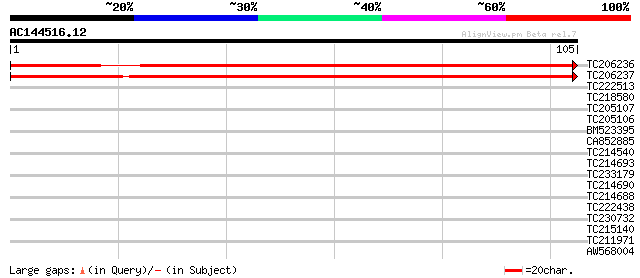
(105 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206236 UP|Q8S8Z5 (Q8S8Z5) Syringolide-induced protein B13-1-1 ... 164 6e-42
TC206237 similar to UP|Q9FMX6 (Q9FMX6) Gb|AAD21732.1, partial (11%) 156 2e-39
TC222513 27 1.2
TC218580 weakly similar to UP|Q7X8Y6 (Q7X8Y6) Tab2 protein, part... 26 2.6
TC205107 similar to UP|Q8H6Q7 (Q8H6Q7) CTV.22, partial (34%) 26 3.4
TC205106 similar to UP|Q8H6Q7 (Q8H6Q7) CTV.22, partial (33%) 26 3.4
BM523395 25 4.5
CA852885 25 4.5
TC214540 similar to UP|Q42768 (Q42768) Acetohydroxyacid synthase... 25 5.8
TC214693 UP|O04664 (O04664) Small Ras-like GTP-binding protein (... 25 5.8
TC233179 25 5.8
TC214690 homologue to UP|Q8S2Z7 (Q8S2Z7) GTP-binding protein, co... 24 10.0
TC214688 homologue to UP|Q8S2Z7 (Q8S2Z7) GTP-binding protein, pa... 24 10.0
TC222438 24 10.0
TC230732 similar to UP|O48796 (O48796) F24O1.5, partial (46%) 24 10.0
TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 24 10.0
TC211971 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-... 24 10.0
AW568004 weakly similar to GP|20196847|gb| expressed protein {Ar... 24 10.0
>TC206236 UP|Q8S8Z5 (Q8S8Z5) Syringolide-induced protein B13-1-1 (Fragment),
complete
Length = 2439
Score = 164 bits (415), Expect = 6e-42
Identities = 79/105 (75%), Positives = 87/105 (82%)
Frame = +1
Query: 1 MGLLWWRKGKNSQPEAKLSTTENAAKNGGASAVKPTTEAPGMNGAVEVKRPKNASVSVFE 60
MGLLWWRKG +Q + + AK+ GA + KP TEAPGMNGAVEV RP N +VS+FE
Sbjct: 2044 MGLLWWRKGNKTQAQPE-------AKSNGADSTKPLTEAPGMNGAVEVPRPSNTTVSIFE 2202
Query: 61 FGSVAASNDKVTLAGYCPVSEDLEPCRWEILPAVESNAPQFRVVF 105
FGSVAASNDKVTLAGYCPVSEDLEPCRWEILPAV+SNAPQFRVVF
Sbjct: 2203 FGSVAASNDKVTLAGYCPVSEDLEPCRWEILPAVQSNAPQFRVVF 2337
>TC206237 similar to UP|Q9FMX6 (Q9FMX6) Gb|AAD21732.1, partial (11%)
Length = 607
Score = 156 bits (394), Expect = 2e-39
Identities = 75/105 (71%), Positives = 86/105 (81%)
Frame = +3
Query: 1 MGLLWWRKGKNSQPEAKLSTTENAAKNGGASAVKPTTEAPGMNGAVEVKRPKNASVSVFE 60
MGLLW +KG QP+ + ++ + K+ G A KP TEAPGMNGAVEV RP N +VS+FE
Sbjct: 165 MGLLW*QKGNKPQPQLQAKSS-SIPKSNGTDATKPLTEAPGMNGAVEVPRPPNTTVSIFE 341
Query: 61 FGSVAASNDKVTLAGYCPVSEDLEPCRWEILPAVESNAPQFRVVF 105
FGSV ASND+VTLAGYCPVSEDLEPCRWEILPAV+SNAPQFRVVF
Sbjct: 342 FGSVTASNDQVTLAGYCPVSEDLEPCRWEILPAVQSNAPQFRVVF 476
>TC222513
Length = 455
Score = 27.3 bits (59), Expect = 1.2
Identities = 9/25 (36%), Positives = 14/25 (56%)
Frame = +2
Query: 5 WWRKGKNSQPEAKLSTTENAAKNGG 29
WWR+G+ S A + T N+ + G
Sbjct: 122 WWRRGRESAETASVDKTRNSGSDTG 196
>TC218580 weakly similar to UP|Q7X8Y6 (Q7X8Y6) Tab2 protein, partial (7%)
Length = 672
Score = 26.2 bits (56), Expect = 2.6
Identities = 15/45 (33%), Positives = 23/45 (50%), Gaps = 3/45 (6%)
Frame = -2
Query: 16 AKLSTTENA---AKNGGASAVKPTTEAPGMNGAVEVKRPKNASVS 57
A ST E + K A A++ T++PG+N + RP N+ S
Sbjct: 374 ASASTAETSNPFIKAANAVALEDATDSPGINVLSSMSRPNNSRSS 240
>TC205107 similar to UP|Q8H6Q7 (Q8H6Q7) CTV.22, partial (34%)
Length = 1762
Score = 25.8 bits (55), Expect = 3.4
Identities = 26/79 (32%), Positives = 35/79 (43%), Gaps = 11/79 (13%)
Frame = +2
Query: 16 AKLSTTENAAKNGGA--SAVKPTTEAP--GMNGAVEVKRPKNASVSVFEFGSVAASNDKV 71
A+ S + A N A S TE P + AV+ K S +V + GSV + ND++
Sbjct: 524 AEFSAPDGAHGNALAATSGKSTVTEQPLERLINAVKSMSSKALSAAVMDIGSVVSMNDRI 703
Query: 72 TLAGYCP-------VSEDL 83
AG P V EDL
Sbjct: 704 --AGSAPGNGSRAAVGEDL 754
>TC205106 similar to UP|Q8H6Q7 (Q8H6Q7) CTV.22, partial (33%)
Length = 1796
Score = 25.8 bits (55), Expect = 3.4
Identities = 18/46 (39%), Positives = 24/46 (52%), Gaps = 7/46 (15%)
Frame = +3
Query: 45 AVEVKRPKNASVSVFEFGSVAASNDKVTLAGYCP-------VSEDL 83
AV+ PK S +V + GSV + ND++ AG P V EDL
Sbjct: 564 AVKSMSPKALSSAVSDIGSVVSMNDRI--AGSAPGNGSRAAVGEDL 695
>BM523395
Length = 430
Score = 25.4 bits (54), Expect = 4.5
Identities = 11/36 (30%), Positives = 19/36 (52%)
Frame = +2
Query: 6 WRKGKNSQPEAKLSTTENAAKNGGASAVKPTTEAPG 41
W++ + ++ + L +TE A K G +KP A G
Sbjct: 308 WQEKREAKRQMYLMSTEKAVKLGERKDLKPAMSAAG 415
>CA852885
Length = 286
Score = 25.4 bits (54), Expect = 4.5
Identities = 20/90 (22%), Positives = 37/90 (40%), Gaps = 2/90 (2%)
Frame = +1
Query: 10 KNSQPEAKLSTTENAAKNGGASAVKPTTEAPGMNGAVEVKRPKNASVSVFEFGSVAASND 69
K Q + L N GG S+V V + +N+++ S +S+D
Sbjct: 4 KKXQTQGHLCVPGNGTSAGGFSSV-----------LVRLCSMQNSAMEA----SKLSSSD 138
Query: 70 KVTLAGYC--PVSEDLEPCRWEILPAVESN 97
+ +C P+S+D+E W + P + +
Sbjct: 139QNVRCHHCAGPLSKDMETSHWTVSPLIRDS 228
>TC214540 similar to UP|Q42768 (Q42768) Acetohydroxyacid synthase , partial
(39%)
Length = 1391
Score = 25.0 bits (53), Expect = 5.8
Identities = 11/35 (31%), Positives = 17/35 (48%)
Frame = +2
Query: 5 WWRKGKNSQPEAKLSTTENAAKNGGASAVKPTTEA 39
WWR+ S+ A L +T A + PTT++
Sbjct: 1253 WWRRVGKSETPATLPSTGPADSPSDTAPPSPTTDS 1357
>TC214693 UP|O04664 (O04664) Small Ras-like GTP-binding protein
(AT5g55190/MCO15_14), partial (79%)
Length = 851
Score = 25.0 bits (53), Expect = 5.8
Identities = 10/24 (41%), Positives = 12/24 (49%)
Frame = +2
Query: 69 DKVTLAGYCPVSEDLEPCRWEILP 92
D + L GYC E P RW + P
Sbjct: 491 DSILLLGYCWAREVWRPQRWILYP 562
>TC233179
Length = 626
Score = 25.0 bits (53), Expect = 5.8
Identities = 11/28 (39%), Positives = 16/28 (56%)
Frame = +3
Query: 14 PEAKLSTTENAAKNGGASAVKPTTEAPG 41
P +K STT + G S+ P T++PG
Sbjct: 531 PSSKPSTTGESRGRGPRSSATPFTDSPG 614
>TC214690 homologue to UP|Q8S2Z7 (Q8S2Z7) GTP-binding protein, complete
Length = 949
Score = 24.3 bits (51), Expect = 10.0
Identities = 10/24 (41%), Positives = 12/24 (49%)
Frame = +1
Query: 69 DKVTLAGYCPVSEDLEPCRWEILP 92
D + L GYC E P RW + P
Sbjct: 301 DSILLLGYCWTGEVWWPQRWILYP 372
>TC214688 homologue to UP|Q8S2Z7 (Q8S2Z7) GTP-binding protein, partial
(84%)
Length = 729
Score = 24.3 bits (51), Expect = 10.0
Identities = 10/24 (41%), Positives = 12/24 (49%)
Frame = +3
Query: 69 DKVTLAGYCPVSEDLEPCRWEILP 92
D + L GYC E P RW + P
Sbjct: 75 DSILLLGYCWTGEIWWPQRWILYP 146
>TC222438
Length = 365
Score = 24.3 bits (51), Expect = 10.0
Identities = 11/38 (28%), Positives = 19/38 (49%)
Frame = +2
Query: 61 FGSVAASNDKVTLAGYCPVSEDLEPCRWEILPAVESNA 98
+G + ++TL G CP+ E C E L + ++A
Sbjct: 17 YGEILFCQKEMTLDGSCPLGTSGESCFLEFLNRLGASA 130
>TC230732 similar to UP|O48796 (O48796) F24O1.5, partial (46%)
Length = 1178
Score = 24.3 bits (51), Expect = 10.0
Identities = 7/13 (53%), Positives = 10/13 (76%)
Frame = +1
Query: 3 LLWWRKGKNSQPE 15
+LWWRK N+ P+
Sbjct: 811 ILWWRKNANNTPK 849
>TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(91%)
Length = 1885
Score = 24.3 bits (51), Expect = 10.0
Identities = 15/35 (42%), Positives = 21/35 (59%)
Frame = -3
Query: 41 GMNGAVEVKRPKNASVSVFEFGSVAASNDKVTLAG 75
G+ AVEV P+NA + FE G A + D+ + AG
Sbjct: 470 GIVVAVEVVEPENAVAAFFE-GKGAVAADEASGAG 369
>TC211971 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-binding
factor 2, partial (20%)
Length = 959
Score = 24.3 bits (51), Expect = 10.0
Identities = 16/43 (37%), Positives = 20/43 (46%)
Frame = +3
Query: 15 EAKLSTTENAAKNGGASAVKPTTEAPGMNGAVEVKRPKNASVS 57
EAKL A GG+S KPT PG + + K S+S
Sbjct: 537 EAKLQ----AISQGGSSHRKPTNSPPGSSNKHNDENGKLKSIS 653
>AW568004 weakly similar to GP|20196847|gb| expressed protein {Arabidopsis
thaliana}, partial (17%)
Length = 436
Score = 24.3 bits (51), Expect = 10.0
Identities = 16/44 (36%), Positives = 20/44 (45%)
Frame = -1
Query: 45 AVEVKRPKNASVSVFEFGSVAASNDKVTLAGYCPVSEDLEPCRW 88
A + +R + SVS V +V A YC V EDL RW
Sbjct: 220 AYDSERVEATSVSAHSNKVVGCGAIRVEHALYCSVGEDLL*RRW 89
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.127 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,493,139
Number of Sequences: 63676
Number of extensions: 42212
Number of successful extensions: 195
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 193
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 193
length of query: 105
length of database: 12,639,632
effective HSP length: 81
effective length of query: 24
effective length of database: 7,481,876
effective search space: 179565024
effective search space used: 179565024
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 51 (24.3 bits)
Medicago: description of AC144516.12


