

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144515.12 - phase: 0
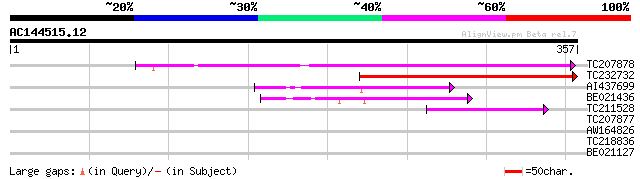
(357 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207878 similar to UP|Q9FI34 (Q9FI34) Emb|CAB68190.1, partial (... 181 3e-46
TC232732 120 7e-28
AI437699 similar to GP|15810016|gb| AT3g58520/F14P22_110 {Arabid... 89 2e-18
BE021436 59 4e-09
TC211528 weakly similar to UP|GAS1_YEAST (P22146) Glycolipid anc... 49 3e-06
TC207877 weakly similar to UP|Q9FI34 (Q9FI34) Emb|CAB68190.1, pa... 40 0.002
AW164826 39 0.003
TC218836 weakly similar to GB|AAM98310.1|22655436|AY142046 At2g1... 29 3.7
BE021127 29 3.7
>TC207878 similar to UP|Q9FI34 (Q9FI34) Emb|CAB68190.1, partial (44%)
Length = 1266
Score = 181 bits (460), Expect = 3e-46
Identities = 103/280 (36%), Positives = 157/280 (55%), Gaps = 3/280 (1%)
Frame = +2
Query: 80 VEKYHHVFMQ--FQPSPGLP-PRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLML 136
+ +Y ++F + F S G P P L+ +AL LH EE+ + + NQ + RL +LLML
Sbjct: 29 IRRYPNIFNESSFLDSGGSPVPCFSLSPEALELHHEEVNI--LQQNQLELRDRLCKLLML 202
Query: 137 AGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRK 196
LP+ I++LKWD+GLP+DY + + +P+ F + D G L+L+ W
Sbjct: 203 TSDRILPLQTIDQLKWDLGLPYDYQHSFVPNHPESFLYVRLPDDRIG-----LKLLFWDD 367
Query: 197 ELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTLPYISPYENA 256
+L++ EL+K D + ++FP+ + F L+++ W++ WQ LPY SPY NA
Sbjct: 368 KLAIPELQKNTSLQQKAEDIKNGSLEFPISFTRGFGLKRKCMEWLKEWQKLPYTSPYINA 547
Query: 257 FHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHHPGIFY 316
HLD +D +EK V + HELL L + K+TE +N+ L L +F K HPGIFY
Sbjct: 548 SHLDPRTDVSEKRIVGVFHELLHLTLHKQTERKNVSNLRRPLALPQKFTKVFERHPGIFY 727
Query: 317 LSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLMTK 356
+S + TQT VLREAY + V+NH ++ R + L+ K
Sbjct: 728 ISKRSDTQTVVLREAYNGQEPVQNHALVQIREEFASLLKK 847
>TC232732
Length = 796
Score = 120 bits (302), Expect = 7e-28
Identities = 58/137 (42%), Positives = 85/137 (61%)
Frame = +3
Query: 221 IQFPMFLPKSFALEKRVKTWVEGWQTLPYISPYENAFHLDSNSDQAEKWTVAILHELLSL 280
I+FP F K L ++++ W+ +Q LPYISPY++ +LD NSD A+K V +LHELLSL
Sbjct: 51 IEFPFFPSKGLRLRRKIENWLNEFQKLPYISPYDDFSNLDPNSDIADKRLVGVLHELLSL 230
Query: 281 LVSKKTESENLICYGECLGLGSRFKKALVHHPGIFYLSNKIRTQTAVLREAYRNEFLVKN 340
V E + L C + GL + +A HP +FYLS + +T T +L+EAY N+ ++
Sbjct: 231 FVEHSAERKKLFCLKKYFGLPQKVHRAFERHPHMFYLSFRNKTCTVILKEAYSNKSAIEK 410
Query: 341 HPVMGTRYWYIHLMTKA 357
HP++ R YI LM K+
Sbjct: 411 HPLLRVRKKYIKLMKKS 461
>AI437699 similar to GP|15810016|gb| AT3g58520/F14P22_110 {Arabidopsis
thaliana}, partial (17%)
Length = 382
Score = 89.4 bits (220), Expect = 2e-18
Identities = 49/128 (38%), Positives = 77/128 (59%), Gaps = 2/128 (1%)
Frame = +3
Query: 155 GLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRKELSVSELEKRVMGFNYDA 214
GLP + TL+ +P++F V+ P V++L+L W EL+VS L+K G +
Sbjct: 3 GLPDTFHKTLVPQFPNHFQF--VKSP---NGVVSLKLSRWPDELAVSALQKHNEGGTHYR 167
Query: 215 DKRRHD--IQFPMFLPKSFALEKRVKTWVEGWQTLPYISPYENAFHLDSNSDQAEKWTVA 272
+ +R + FPM P+ + + +V+TW+E +Q LPY+SPY ++ +D NSD EK V
Sbjct: 168 EFKRGQSALAFPMRFPRGYGAQTKVRTWMEEFQKLPYVSPYVDSTKIDPNSDLMEKRVVG 347
Query: 273 ILHELLSL 280
+LHE+LSL
Sbjct: 348 VLHEILSL 371
>BE021436
Length = 427
Score = 58.5 bits (140), Expect = 4e-09
Identities = 46/148 (31%), Positives = 66/148 (44%), Gaps = 15/148 (10%)
Frame = +2
Query: 159 DYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRKELSVSELEKR-----VMGFNYD 213
D+ L+ YP +F V G++ LALE W L+++ E R V+ D
Sbjct: 2 DFKDCLVPKYPQFFAV----RRFRGRDSLALE--DWDSTLALTARESRLAQEGVVNLKAD 163
Query: 214 ADKRRHDIQ----------FPMFLPKSFALEKRVKTWVEGWQTLPYISPYENAFHLDSNS 263
++R+ I F M P F +E WQ L + SPY NA D+
Sbjct: 164 GNRRKVKISRDGNYLGPFAFKMNFPAGFRPNVGYLEQLERWQKLEFPSPYLNARRFDAAD 343
Query: 264 DQAEKWTVAILHELLSLLVSKKTESENL 291
+A K VA++HELLSL + K+ S L
Sbjct: 344 PKARKRAVAVIHELLSLTMEKRMTSLQL 427
>TC211528 weakly similar to UP|GAS1_YEAST (P22146) Glycolipid anchored
surface protein precursor (Glycoprotein GP115), partial
(6%)
Length = 548
Score = 48.9 bits (115), Expect = 3e-06
Identities = 25/77 (32%), Positives = 42/77 (54%)
Frame = +3
Query: 263 SDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHHPGIFYLSNKIR 322
S + EK ++HE+LSL + K+T ++ + E + + L+ HP +FY+S K
Sbjct: 9 SREKEKHACGVVHEILSLTLEKRTLVDHFTHFREEFRFSQQLRGMLIRHPDMFYVSLKGD 188
Query: 323 TQTAVLREAYRNEFLVK 339
+ LRE YR+ LV+
Sbjct: 189 RDSVFLREGYRDSQLVE 239
>TC207877 weakly similar to UP|Q9FI34 (Q9FI34) Emb|CAB68190.1, partial (8%)
Length = 380
Score = 40.0 bits (92), Expect = 0.002
Identities = 19/46 (41%), Positives = 29/46 (62%)
Frame = +2
Query: 115 EVHNSRTNQEDAAQRLARLLMLAGMARLPIYVIEKLKWDMGLPHDY 160
E++ + NQ + L +LLMLA LP+ I++LKWD+G +DY
Sbjct: 2 EMNILQQNQWELQVMLCKLLMLASDRILPLQTIDQLKWDLGXXYDY 139
>AW164826
Length = 414
Score = 39.3 bits (90), Expect = 0.003
Identities = 33/127 (25%), Positives = 57/127 (43%), Gaps = 1/127 (0%)
Frame = +3
Query: 47 KNHIVSSPSKSLSLYNASMLKASLNLPMITI-KLVEKYHHVFMQFQPSPGLPPRIKLTAQ 105
+N I+S PS+SL + L V K+ HVF F+ +LT +
Sbjct: 6 QNLILSEPSQSLPISRLETLARRFGFTRHESGAFVLKFPHVFEIFEHPVQRILFCRLTRK 185
Query: 106 ALALHKEEMEVHNSRTNQEDAAQRLARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLL 165
A+ ++E + + A RL +LLM++ RL + + + GLP D+ +++
Sbjct: 186 AILQIEQERRALAAELPR--AVTRLRKLLMMSNHGRLRLEHVRIARAAYGLPDDFENSVV 359
Query: 166 AYYPDYF 172
YP +F
Sbjct: 360 LRYPQFF 380
>TC218836 weakly similar to GB|AAM98310.1|22655436|AY142046
At2g19080/T20K24.9 {Arabidopsis thaliana;} , partial
(38%)
Length = 713
Score = 28.9 bits (63), Expect = 3.7
Identities = 20/100 (20%), Positives = 45/100 (45%), Gaps = 5/100 (5%)
Frame = +3
Query: 59 SLYNASMLKASLNLPMITIKLVEKYHHVFMQFQPSPGLPPRIKLTAQALA-----LHKEE 113
+L +SML+ + ++ V++ F++ PSP PR A + A K +
Sbjct: 189 ALPESSMLRIKFSEHAHLVRYVQQCKTEFIEAAPSPSSAPRFHTGASSSASKSHSSSKPK 368
Query: 114 MEVHNSRTNQEDAAQRLARLLMLAGMARLPIYVIEKLKWD 153
+ +T +E +R A+ ++A + + +++ +D
Sbjct: 369 SKPKREKTQEEKTFKRKAKYFVVAQLVAVVVFLTLMTSFD 488
>BE021127
Length = 378
Score = 28.9 bits (63), Expect = 3.7
Identities = 23/64 (35%), Positives = 34/64 (52%), Gaps = 2/64 (3%)
Frame = +3
Query: 231 FALEKRVKTWVEGWQTLPYISPYENAFHLDSNSDQAEKWTVAIL--HELLSLLVSKKTES 288
F E+ +K ++ + TL SP+ NA+HL S S A W I+ H++LS S+
Sbjct: 81 FGDEENIK-YMGFFTTLSRNSPFINAYHLGSLSVSAV*WHKLIVTQHKILSRRFSEMILV 257
Query: 289 ENLI 292
NLI
Sbjct: 258 TNLI 269
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,388,403
Number of Sequences: 63676
Number of extensions: 247285
Number of successful extensions: 1202
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1198
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1201
length of query: 357
length of database: 12,639,632
effective HSP length: 98
effective length of query: 259
effective length of database: 6,399,384
effective search space: 1657440456
effective search space used: 1657440456
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144515.12


