

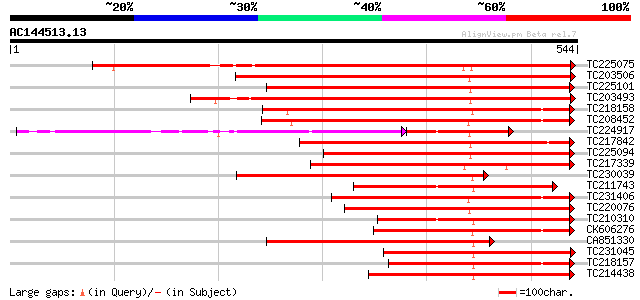
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.13 + phase: 0
(544 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 441 e-124
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 383 e-106
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 379 e-105
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 377 e-105
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 353 8e-98
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 352 2e-97
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 214 6e-91
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 311 6e-85
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 310 1e-84
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 280 1e-75
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 274 8e-74
TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 261 7e-70
TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (... 255 3e-68
TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 pr... 246 2e-65
TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 244 9e-65
CK606276 238 5e-63
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 230 1e-60
TC231045 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase iso... 218 5e-57
TC218157 similar to UP|Q8GUF8 (Q8GUF8) At2g47550/T30B22.15, part... 217 9e-57
TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (F... 214 6e-56
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 441 bits (1133), Expect = e-124
Identities = 230/478 (48%), Positives = 309/478 (64%), Gaps = 14/478 (2%)
Frame = +1
Query: 80 TINEAIGAINNMTKI-STFS---VNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAG- 134
++ E I AI + I S F N R AI DC +LLD S L+W+L + +
Sbjct: 160 SVTEVIAAIRQLASILSRFGSPLANFRLSTAIADCLDLLDLSSDVLSWALSASQNPKGKH 339
Query: 135 DRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQL 194
+ T +L WLSAAL++ +TC+EGFEGT+ ++ +S + QV L+ +L+
Sbjct: 340 NSTGNLSSDLRTWLSAALAHPETCMEGFEGTNSIVKGLVSAGIGQVVSLVEQLLAQVL-- 513
Query: 195 NRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTI 254
P S+ +FP W+ +++LL++ D VALDGSG Y I
Sbjct: 514 -------PAQDQFDAASSKG--QFPSWIKPKERKLLQAI--AVTPDVTVALDGSGNYAKI 660
Query: 255 NEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWT 314
+AV AAP +S +R VI VKKG+Y EN+++KKK NIM++G G+ T+++ NR+ + GWT
Sbjct: 661 MDAVLAAPDYSMKRFVILVKKGVYVENVEIKKKKWNIMILGQGMDATVISGNRSVVDGWT 840
Query: 315 TFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHS 374
TFR+ATFAVSG+GFIA+D++F+NTAGP HQAVALR DSD S FFRC I G QD+LY H+
Sbjct: 841 TFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHT 1020
Query: 375 LRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQ 434
+RQF+R+C I GT+D+IFG+ AV QNC + + LP QK TITA GRK P++ TGF+ Q
Sbjct: 1021MRQFFRDCTISGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHGRKDPNEPTGFSFQ 1200
Query: 435 ------DSYVLAS---QPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLW 485
DS ++ S TYLGRPWK YSRTV++ +YMS ++ GWLEW GNFALDTL+
Sbjct: 1201FCNITADSDLIPSVGTAQTYLGRPWKSYSRTVFMQSYMSEVIGAEGWLEWNGNFALDTLY 1380
Query: 486 YGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
Y EY N G G+ +A RVKWPGYH + D+S A FTV +F+ G WLP TGV FTAGL+
Sbjct: 1381YAEYMNTGAGAGVANRVKWPGYHALNDSSQASNFTVSQFIEGNLWLPSTGVTFTAGLT 1554
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 383 bits (984), Expect = e-106
Identities = 181/337 (53%), Positives = 235/337 (69%), Gaps = 10/337 (2%)
Frame = +2
Query: 217 EFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKG 276
+FP W+ + D+ LL AD V+ G+G +TI EA+ AP HS RR +IYV+ G
Sbjct: 8 KFPGWLNKRDRRLLSLPLSAIQADITVSKTGNGTVKTIAEAIKKAPEHSTRRFIIYVRAG 187
Query: 277 LYKEN-IDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTF 335
Y+EN + + +K TN+M +GDG G+T++T +N + G TTF +A+FA SG GFIA+D+TF
Sbjct: 188 RYEENNLKVGRKKTNVMFIGDGKGKTVITGKKNVIDGMTTFHSASFAASGAGFIARDITF 367
Query: 336 RNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNG 395
N AGP HQAVALRV +D + +RC+I G QD+ Y HS RQF+REC IYGT+DFIFGN
Sbjct: 368 ENYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFIFGNA 547
Query: 396 AAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVL---------ASQPTYL 446
A V Q C IY R P+ QK TITAQ RK P+Q+TG ++ + +L S PTYL
Sbjct: 548 AVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISLHNCRILPAPDLAPVKGSFPTYL 727
Query: 447 GRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPG 506
GRPWK+YSRTVY+ +YM + PRGWLEW G+FAL+TL+YGEY NYGPG+++ RV+WPG
Sbjct: 728 GRPWKQYSRTVYLVSYMGDHIHPRGWLEWNGDFALNTLYYGEYMNYGPGAAVGQRVQWPG 907
Query: 507 YHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
Y VIK A FTV +F++G +WLP TGV F AGLS
Sbjct: 908 YRVIKSTMEANRFTVAQFISGSAWLPSTGVAFAAGLS 1018
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 379 bits (972), Expect = e-105
Identities = 179/308 (58%), Positives = 231/308 (74%), Gaps = 12/308 (3%)
Frame = +3
Query: 247 GSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSN 306
G Y + +AV AAP++S +R+VI++K+G+Y EN+++KKK N+MMVGDG+ TI++ N
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGDGMDATIISGN 386
Query: 307 RNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGN 366
R+F+ GWTTFR+ATFAVSG+GFIA+D+TF+NTAGP HQAVALR DSD S FFRC I G
Sbjct: 387 RSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGY 566
Query: 367 QDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPH 426
QD+LY H++RQFYREC+I GT+DFIFG+ A+ QNC I + LP QK TITA GRK+P
Sbjct: 567 QDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAHGRKNPD 746
Query: 427 QSTGFTIQDSYVLA------------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLE 474
+ TGF+IQ + A S TYLGRPWK YSRT+++ +Y+S +++P GWLE
Sbjct: 747 EPTGFSIQFCNISADYDLVNSVNSCNSTHTYLGRPWKPYSRTIFMQSYISDVLRPEGWLE 926
Query: 475 WLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRT 534
W G+FALDTL+Y EY NYGPG+ +A RVKW GYHV+ D+S A FTV +F+ G LP T
Sbjct: 927 WNGDFALDTLYYAEYMNYGPGAGVANRVKWQGYHVMNDSSQASNFTVSQFIEGNLCLPST 1106
Query: 535 GVKFTAGL 542
GV FTAGL
Sbjct: 1107GVTFTAGL 1130
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 377 bits (968), Expect = e-105
Identities = 193/388 (49%), Positives = 255/388 (64%), Gaps = 18/388 (4%)
Frame = +1
Query: 174 SGSVTQVTQLISNVLSLYTQLNR------LPFRPPRNTTLHETSTDESLEFPEWMTEADQ 227
S ++ +++L+SN L++++ +P + R FP W+ D+
Sbjct: 4 SNNLKDLSELVSNCLAIFSGAGAGDDFAGVPIQNRRRLMAMREDN-----FPTWLNGRDR 168
Query: 228 ELLKSKPHGKI-ADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKE-NIDMK 285
LL S P +I AD VV+ DG+G +TI EA+ P +S+RR +IY++ G Y+E N+ +
Sbjct: 169 RLL-SLPLSQIQADIVVSKDGNGTVKTIAEAIKKVPEYSSRRIIIYIRAGRYEEDNLKLG 345
Query: 286 KKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQ 345
+K TN+M +GDG G+T++T RN+ Q TTF TA+FA SG GFIAKDMTF N AGP HQ
Sbjct: 346 RKKTNVMFIGDGKGKTVITGGRNYYQNLTTFHTASFAASGSGFIAKDMTFENYAGPGRHQ 525
Query: 346 AVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIY 405
AVALRV +D + +RC+I G QDT+Y HS RQFYREC+IYGT+DFIFGN A V QNC ++
Sbjct: 526 AVALRVGADHAVVYRCNIIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCTLW 705
Query: 406 TRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA---------SQPTYLGRPWKEYSRT 456
R P+ QK TITAQ RK P+Q+TG +I + ++A S PTYLGRPWK Y+RT
Sbjct: 706 ARKPMAQQKNTITAQNRKDPNQNTGISIHNCRIMATPDLEASKGSYPTYLGRPWKLYART 885
Query: 457 VYINTYMSSMVQPRGWLEW-LGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASA 515
V++ +Y+ V PRGWLEW +FALDT +YGEY NYGPGS+L RV W GY I
Sbjct: 886 VFMLSYIGDHVHPRGWLEWNTSSFALDTCYYGEYMNYGPGSALGQRVNWAGYRAINSTVE 1065
Query: 516 AGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
A FTV +F++G SWLP TGV F AGLS
Sbjct: 1066ASRFTVGQFISGSSWLPSTGVAFIAGLS 1149
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 353 bits (907), Expect = 8e-98
Identities = 175/311 (56%), Positives = 217/311 (69%), Gaps = 11/311 (3%)
Frame = +3
Query: 243 VALDGSGQYRTINEAVNAAPSHS---NRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIG 299
V+ DGSG + TIN+A+ AAP+ S + +IYV G+Y+EN+ + KK T +MMVGDGI
Sbjct: 159 VSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAGVYEENVSIDKKKTYLMMVGDGIN 338
Query: 300 QTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFF 359
+TI+T NR+ + GWTTF +AT AV G+GF+ +MT RNTAG V HQAVALR +D S F+
Sbjct: 339 KTIITGNRSVVDGWTTFSSATLAVVGQGFVGVNMTIRNTAGAVKHQAVALRSGADLSTFY 518
Query: 360 RCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITA 419
CS EG QDTLY HSLRQFY EC+IYGT+DFIFGN V QNCK+Y R+P+ Q ITA
Sbjct: 519 SCSFEGYQDTLYVHSLRQFYSECDIYGTVDFIFGNAKVVFQNCKMYPRLPMSGQFNAITA 698
Query: 420 QGRKSPHQSTGFTIQDSYVLASQ--------PTYLGRPWKEYSRTVYINTYMSSMVQPRG 471
QGR P+Q TG +I + + A+ TYLGRPWKEYSRTVY+ T M S++ +G
Sbjct: 699 QGRTDPNQDTGISIHNCTIRAADDLAASNGVATYLGRPWKEYSRTVYMQTVMDSVIHAKG 878
Query: 472 WLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWL 531
W EW G+FAL TL+Y EY N GPGS RV WPGYHVI +A+ A FTV FL G WL
Sbjct: 879 WREWDGDFALSTLYYAEYSNSGPGSGTDNRVTWPGYHVI-NATDAANFTVSNFLLGDDWL 1055
Query: 532 PRTGVKFTAGL 542
P+TGV +T L
Sbjct: 1056PQTGVSYTNNL 1088
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 352 bits (903), Expect = 2e-97
Identities = 169/313 (53%), Positives = 225/313 (70%), Gaps = 12/313 (3%)
Frame = +1
Query: 242 VVALDGSGQYRTINEAVNAAPSHSNRRH---VIYVKKGLYKENIDMKKKMTNIMMVGDGI 298
+V+ G Y +I +A+ AAP+++ ++YV++GLY+E + + K+ NI++VGDGI
Sbjct: 295 IVSHYGIDNYTSIGDAIAAAPNNTKPEDGYFLVYVREGLYEEYVVIPKEKKNILLVGDGI 474
Query: 299 GQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAF 358
+TI+T N + + GWTTF ++TFAVSG+ FIA D+TFRNTAGP HQAVA+R ++D S F
Sbjct: 475 NKTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDVTFRNTAGPEKHQAVAVRNNADLSTF 654
Query: 359 FRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT 418
+RCS EG QDTLY HSLRQFYRECEIYGT+DFIFGN A V Q CKIY R PLP QK +T
Sbjct: 655 YRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAAVVFQGCKIYARKPLPNQKNAVT 834
Query: 419 AQGRKSPHQSTGFTIQDSYV---------LASQPTYLGRPWKEYSRTVYINTYMSSMVQP 469
AQGR P+Q+TG +IQ+ + L S ++LGRPWK YSRTVY+ +Y+ +++QP
Sbjct: 835 AQGRTDPNQNTGISIQNCSIDAAPDLVADLNSTMSFLGRPWKVYSRTVYLQSYIGNVIQP 1014
Query: 470 RGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGS 529
GWLEW G LDTL+YGE+ NYGPGS+ + RV WPGY ++ +A+ A FTV F G +
Sbjct: 1015AGWLEWNGTVGLDTLFYGEFNNYGPGSNTSNRVTWPGYSLL-NATQAWNFTVLNFTLGNT 1191
Query: 530 WLPRTGVKFTAGL 542
WLP T + +T GL
Sbjct: 1192WLPDTDIPYTEGL 1230
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 214 bits (545), Expect(2) = 6e-91
Identities = 143/377 (37%), Positives = 201/377 (52%), Gaps = 3/377 (0%)
Frame = +2
Query: 7 FLFILILLPSFDQVLSFPDEIPSDIQTQDMQALIAQACMDIENQNSCLTNIHNELTRTGP 66
FL L+L P I S +D+Q+ C C + N
Sbjct: 131 FLMTLLLAPFLFS------SIASSYSFKDIQSW----CNQTPYPQPCEYYLTNHAFNKPI 280
Query: 67 PSPTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLG 126
S + + +L+ + A + N + N E+ A DC +L ++++ L ++
Sbjct: 281 KSKSDFLKVSLQLALERAQRSELNTHALGPKCRNVHEKAAWADCLQLYEYTIQRLNKTIN 460
Query: 127 EMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQ-VTQLIS 185
+ D + WLS AL+N +TC GF + + Y+ ++ VT+L+S
Sbjct: 461 PNTKCNQTDT--------QTWLSTALTNLETCKNGFY--ELGVPDYVLPLMSNNVTKLLS 610
Query: 186 NVLSLYTQLNRLPF--RPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVV 243
N LSL N+ P+ +PP S E FP W+ D++LL+S A+ VV
Sbjct: 611 NTLSL----NKGPYQYKPP--------SYKEG--FPTWVKPGDRKLLQSSSVASNANVVV 748
Query: 244 ALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIV 303
A DGSG+Y T+ AV+AAP S+ R+VIYVK G+Y E +++K NIM+VGDGIG+TI+
Sbjct: 749 AKDGSGKYTTVKAAVDAAPKSSSGRYVIYVKSGVYNEQVEVKGN--NIMLVGDGIGKTII 922
Query: 304 TSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSI 363
T +++ G TTFR+AT A G GFIA+D+TFRNTAG NHQAVA R SD S F+RCS
Sbjct: 923 TGSKSVGGGTTTFRSATVAAVGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYRCSF 1102
Query: 364 EGNQDTLYAHSLRQFYR 380
EG QDTLY HS RQFYR
Sbjct: 1103EGFQDTLYVHSERQFYR 1153
Score = 139 bits (349), Expect(2) = 6e-91
Identities = 64/110 (58%), Positives = 82/110 (74%), Gaps = 7/110 (6%)
Frame = +3
Query: 381 ECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVL- 439
EC+IYGT+DFIFGN AAVLQNC IY R P P + +T+TAQGR P+Q+TG I +S V
Sbjct: 1155 ECDIYGTVDFIFGNAAAVLQNCNIYARTP-PQRTITVTAQGRTDPNQNTGIIIHNSKVTG 1331
Query: 440 ------ASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDT 483
+S +YLGRPW++YSRTV++ TY+ S++ P GW+EW GNFALDT
Sbjct: 1332 ASGFNPSSVKSYLGRPWQKYSRTVFMKTYLDSLINPAGWMEWDGNFALDT 1481
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 311 bits (796), Expect = 6e-85
Identities = 150/273 (54%), Positives = 190/273 (68%), Gaps = 9/273 (3%)
Frame = +3
Query: 279 KENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNT 338
KENID+K+ + N+M+VGDG+G TIVT N N + G TTFR+ATFAV G GFIA+D+TF NT
Sbjct: 6 KENIDIKRTVKNLMIVGDGMGATIVTGNHNAIDGSTTFRSATFAVDGDGFIARDITFENT 185
Query: 339 AGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAV 398
AGP HQAVALR +D S F+RCS G QDTLY ++ RQFYR+C+IYGT+DFIFG+ AV
Sbjct: 186 AGPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDAVAV 365
Query: 399 LQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA---------SQPTYLGRP 449
LQNC IY R P+ Q+ T+TAQGR P+++TG I + + A S T+LGRP
Sbjct: 366 LQNCNIYVRKPMSNQQNTVTAQGRTDPNENTGIIIHNCRITAAGDLKAVQGSFRTFLGRP 545
Query: 450 WKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHV 509
W++YSRTV + + + ++ P GW W GNFAL TL+Y E+ N G G+S GRV W G+ V
Sbjct: 546 WQKYSRTVVMKSALDGLISPAGWFPWSGNFALSTLYYAEHANTGAGASTGGRVDWAGFRV 725
Query: 510 IKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
I A FTV FL GGSW+P +GV F GL
Sbjct: 726 ISSTEAV-KFTVGNFLAGGSWIPGSGVPFDEGL 821
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 310 bits (794), Expect = 1e-84
Identities = 145/250 (58%), Positives = 184/250 (73%), Gaps = 9/250 (3%)
Frame = +3
Query: 302 IVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRC 361
I++ NR+ + GWTTFR+ATFAVSG+GFIA+D++F+NTAGP HQAVALR D+D S FFRC
Sbjct: 3 IISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRC 182
Query: 362 SIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQG 421
I G QD+LY H++RQF+REC I GT+D+IFG+ AV QNC + + LP QK TITA G
Sbjct: 183 GIFGYQDSLYTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHG 362
Query: 422 RKSPHQSTGFTIQDSYVLA---------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGW 472
RK P++ TGF+ Q + A S +YLGRPWK YSRTV++ +YMS +++ GW
Sbjct: 363 RKDPNEPTGFSFQFCNITADSDLVPWVSSTQSYLGRPWKSYSRTVFMQSYMSEVIRGEGW 542
Query: 473 LEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLP 532
LEW GNFAL+TL+YGEY N G G+ LA RVKWPGYH D++ A FTV +F+ G WLP
Sbjct: 543 LEWNGNFALETLYYGEYMNTGAGAGLANRVKWPGYHPFNDSNQASNFTVAQFIEGNLWLP 722
Query: 533 RTGVKFTAGL 542
TGV +TAGL
Sbjct: 723 STGVTYTAGL 752
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 280 bits (716), Expect = 1e-75
Identities = 140/265 (52%), Positives = 174/265 (64%), Gaps = 11/265 (4%)
Frame = +1
Query: 289 TNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVA 348
TN+M+VGDG T++T N NF+ G TTF+TAT A G GFIA+D+ F+NTAGP HQAVA
Sbjct: 7 TNVMLVGDGKDATVITGNLNFIDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQAVA 186
Query: 349 LRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRV 408
LRV +DQS RC I+ QDTLYAHS RQFYR+ I GT+DFIFGN A V Q C + R
Sbjct: 187 LRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVARK 366
Query: 409 PLPLQKVTITAQGRKSPHQSTGFTIQD---------SYVLASQPTYLGRPWKEYSRTVYI 459
P+ Q +TAQGR+ P+Q+TG +IQ V+ S T+LGRPWK+YSRTV +
Sbjct: 367 PMDKQNNMVTAQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTFLGRPWKKYSRTVVM 546
Query: 460 NTYMSSMVQPRGWLEW--LGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAG 517
+ + S + P GW EW L TL+YGEY N GPG+ + RV WPGYH+IK A+ A
Sbjct: 547 QSTLDSHIDPTGWAEWDAQSKDFLQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAAEAS 726
Query: 518 YFTVQRFLNGGSWLPRTGVKFTAGL 542
FTV + + G WL TGV F GL
Sbjct: 727 KFTVAQLIQGNVWLKNTGVNFIEGL 801
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 274 bits (700), Expect = 8e-74
Identities = 134/251 (53%), Positives = 178/251 (70%), Gaps = 9/251 (3%)
Frame = +1
Query: 218 FPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGL 277
FP W++ D++LL++ + + +VA DG+G + TI EAV AP+ S R VI++K G
Sbjct: 160 FPTWLSTKDRKLLQAAVNETNFNLLVAKDGTGNFTTIAEAVAVAPNSSATRFVIHIKAGA 339
Query: 278 YKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRN 337
Y EN+++ +K TN+M VGDGIG+T+V ++RN + GWTTF++AT AV G GFIAK +TF N
Sbjct: 340 YFENVEVIRKKTNLMFVGDGIGKTVVKASRNVVDGWTTFQSATVAVVGDGFIAKGITFEN 519
Query: 338 TAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAA 397
+AGP HQAVALR SD SAF++CS QDTLY HSLRQFYR+C++YGT+DFIFGN A
Sbjct: 520 SAGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDFIFGNAAT 699
Query: 398 VLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQ---------PTYLGR 448
VLQNC +Y R P Q+ TAQGR+ P+Q+TG +I + V A+ YLGR
Sbjct: 700 VLQNCNLYARKPNENQRNLFTAQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLGR 879
Query: 449 PWKEYSRTVYI 459
PWK+YSRTVY+
Sbjct: 880 PWKKYSRTVYL 912
>TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (39%)
Length = 601
Score = 261 bits (666), Expect = 7e-70
Identities = 127/200 (63%), Positives = 148/200 (73%), Gaps = 5/200 (2%)
Frame = +2
Query: 331 KDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDF 390
+D+TFRNTAG NHQAVALR SD S F+RCS EG QDTLY +S RQFYREC+IYGT+DF
Sbjct: 2 QDITFRNTAGATNHQAVALRSGSDLSVFYRCSFEGYQDTLYVYSDRQFYRECDIYGTVDF 181
Query: 391 IFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQP-----TY 445
IFGN A V QNC IY R P P + TITAQGR P+Q+TG +I +S V A+ TY
Sbjct: 182 IFGNAAVVFQNCNIYARNP-PNKVNTITAQGRTDPNQNTGISIHNSKVTAASDLMGVRTY 358
Query: 446 LGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWP 505
LGRPW++YSRTV++ TY+ S++ P GWLEW GNFAL TL+YGEY N GPGSS A RV W
Sbjct: 359 LGRPWQQYSRTVFMKTYLDSLINPEGWLEWSGNFALSTLYYGEYMNTGPGSSTANRVNWL 538
Query: 506 GYHVIKDASAAGYFTVQRFL 525
GYHVI AS A FTV F+
Sbjct: 539 GYHVITSASEASKFTVGNFI 598
>TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (35%)
Length = 851
Score = 255 bits (652), Expect = 3e-68
Identities = 125/239 (52%), Positives = 160/239 (66%), Gaps = 5/239 (2%)
Frame = +2
Query: 309 FMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQD 368
F+ G TF TATFAV G+ FIA+DM FRNTAGP QAVAL +DQ+ ++RC I+ QD
Sbjct: 2 FVDGTPTFSTATFAVFGRNFIARDMGFRNTAGPQKQQAVALMTSADQAVYYRCQIDAFQD 181
Query: 369 TLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQS 428
+LYAHS RQFYREC IYGT+DFIFGN A VLQNC I RVP+ Q+ ITAQG+ P+ +
Sbjct: 182 SLYAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNIMPRVPMQGQQNPITAQGKTDPNMN 361
Query: 429 TGFTIQDSYV-----LASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDT 483
TG +IQ+ + L+S TYLGRPWK YS V++ + M + P GWL W+GN A DT
Sbjct: 362 TGISIQNCNITPFGDLSSVKTYLGRPWKNYSTPVFM*SPMGIFIHPNGWLPWVGNSAPDT 541
Query: 484 LWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
++Y E++N GPG+S RV W G VI A FTV+ FL+G W+ +G F + +
Sbjct: 542 IFYAEFQNVGPGASTKNRVNWKGLRVI-TRKQASMFTVKAFLSGERWITASGAPFKSSI 715
>TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (39%)
Length = 808
Score = 246 bits (628), Expect = 2e-65
Identities = 125/227 (55%), Positives = 152/227 (66%), Gaps = 6/227 (2%)
Frame = +2
Query: 322 AVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRE 381
AV GKGFIAKD+ F N AG HQAVA R SD+S FFRCS G QDTLYAHS RQFYR+
Sbjct: 80 AVKGKGFIAKDIGFVNNAGASKHQAVAFRSGSDRSVFFRCSFNGFQDTLYAHSNRQFYRD 259
Query: 382 CEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVL-- 439
C+I GTIDFIFGN AAV QNCKI R PLP Q TITAQG+K +Q+TG IQ S
Sbjct: 260 CDITGTIDFIFGNAAAVFQNCKIMPRQPLPNQFNTITAQGKKDRNQNTGIIIQKSKFTPL 439
Query: 440 ---ASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNF-ALDTLWYGEYRNYGPG 495
+ PTYLGRPWK++S TV + + + S ++P GW+ W+ N + T++Y EY+N GPG
Sbjct: 440 ENNLTAPTYLGRPWKDFSTTVIMQSDIGSFLKPVGWMSWVPNVEPVSTIFYAEYQNTGPG 619
Query: 496 SSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
+ ++ RVKW GY AG FTVQ F+ G WLP V+F + L
Sbjct: 620 ADVSQRVKWAGYKPTLTDGEAGKFTVQSFIQGPEWLPNAAVQFDSTL 760
>TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (38%)
Length = 678
Score = 244 bits (622), Expect = 9e-65
Identities = 118/198 (59%), Positives = 139/198 (69%), Gaps = 9/198 (4%)
Frame = +1
Query: 354 DQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQ 413
D S F++CS EG QDTLY HS RQFYREC IYGT+DFIFGN A VLQNC I+ R P P +
Sbjct: 1 DLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNP-PNK 177
Query: 414 KVTITAQGRKSPHQSTGFTIQDSYVLASQP---------TYLGRPWKEYSRTVYINTYMS 464
TITAQGR P+Q+TG +I +S V A+ TYLGRPWK+YSRTV++ TY+
Sbjct: 178 VNTITAQGRTDPNQNTGISIHNSRVTAASDLRPVQNSVRTYLGRPWKQYSRTVFMKTYLD 357
Query: 465 SMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRF 524
++ P GW+EW GNFALDTL+YGEY N GPGSS A RVKW GY VI AS A F+V F
Sbjct: 358 GLINPAGWMEWSGNFALDTLYYGEYMNTGPGSSTARRVKWSGYRVITSASEASKFSVANF 537
Query: 525 LNGGSWLPRTGVKFTAGL 542
+ G +WLP T V FT L
Sbjct: 538 IAGNAWLPSTKVPFTPSL 591
>CK606276
Length = 751
Score = 238 bits (607), Expect = 5e-63
Identities = 110/205 (53%), Positives = 144/205 (69%), Gaps = 12/205 (5%)
Frame = +1
Query: 350 RVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVP 409
R +D S F+ CS EG QDTLY HSLRQFY+ C+IYGT+DFIFGN AA+LQ+C +Y R+P
Sbjct: 1 RNGADMSTFYNCSFEGYQDTLYVHSLRQFYKSCDIYGTVDFIFGNAAALLQDCNMYPRLP 180
Query: 410 LPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQ------------PTYLGRPWKEYSRTV 457
+ Q ITAQGR P+Q+TG +IQ+ ++A+ TYLGRPWKEYSRTV
Sbjct: 181 MQNQFNAITAQGRTDPNQNTGISIQNCCIIAASDLGDATNNYNGIKTYLGRPWKEYSRTV 360
Query: 458 YINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAG 517
Y+ +++ ++ P+GW EW G+FAL TL+Y E+ N+GPGS+ + RV W GYH+I D A
Sbjct: 361 YMQSFIDGLIDPKGWNEWSGDFALSTLYYAEFANWGPGSNTSNRVAWEGYHLI-DEKDAD 537
Query: 518 YFTVQRFLNGGSWLPRTGVKFTAGL 542
FTV +F+ G WLP+TGV F AGL
Sbjct: 538 DFTVHKFIQGEKWLPQTGVPFXAGL 612
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 230 bits (586), Expect = 1e-60
Identities = 117/229 (51%), Positives = 156/229 (68%), Gaps = 10/229 (4%)
Frame = +2
Query: 247 GSGQYRTINEAVNAAPSHSNR-RHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTS 305
GSG ++T+ +A+NAA + R VI+VKKG+Y+ENI++ NIM+VGDG+ TI+TS
Sbjct: 14 GSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYRENIEVALHNDNIMLVGDGLRNTIITS 193
Query: 306 NRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEG 365
R+ G+TT+ +AT + G FIA+D+TF+N+AG QAVALR SD S F+RC I G
Sbjct: 194 ARSVQDGYTTYSSATAGIDGLHFIARDITFQNSAGVHKGQAVALRSASDLSVFYRCGIMG 373
Query: 366 NQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSP 425
QDTL AH+ RQFYR+C IYGT+DFIFGN A V QNC I+ R PL Q ITAQGR P
Sbjct: 374 YQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVFQNCYIFARRPLEGQANMITAQGRGDP 553
Query: 426 HQSTGFTIQDSYVLASQP---------TYLGRPWKEYSRTVYINTYMSS 465
Q+TG +I +S + A+ T+LGRPW++YSR V + T+M +
Sbjct: 554 FQNTGISIHNSQIRAAPDLKPVVDKYNTFLGRPWQQYSRVVVMKTFMDT 700
>TC231045 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (69%)
Length = 747
Score = 218 bits (555), Expect = 5e-57
Identities = 99/194 (51%), Positives = 128/194 (65%), Gaps = 9/194 (4%)
Frame = +2
Query: 359 FRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT 418
++C QD HS RQFYR+C++YGT+DF GN A VLQNC +Y R P Q+ T
Sbjct: 2 YKCXFVXYQDXXXVHSXRQFYRDCDVYGTVDFXXGNAATVLQNCNLYARKPNENQRNLFT 181
Query: 419 AQGRKSPHQSTGFTIQDSYVLASQP---------TYLGRPWKEYSRTVYINTYMSSMVQP 469
AQGR+ P+Q+TG +I + V A+ YLGRPWK+YSRTVY+N+YM ++ P
Sbjct: 182 AQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLGRPWKKYSRTVYLNSYMEDLIDP 361
Query: 470 RGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGS 529
+GWLEW G FALDTL+YGEY N GPGS+ + RV WPGY VIK+A+ A FTV+ F+ G
Sbjct: 362 KGWLEWNGTFALDTLYYGEYNNRGPGSNTSARVTWPGYRVIKNATEANQFTVRNFIQGNE 541
Query: 530 WLPRTGVKFTAGLS 543
WL T + F + S
Sbjct: 542 WLSSTDIPFFSDFS 583
>TC218157 similar to UP|Q8GUF8 (Q8GUF8) At2g47550/T30B22.15, partial (49%)
Length = 748
Score = 217 bits (553), Expect = 9e-57
Identities = 106/187 (56%), Positives = 126/187 (66%), Gaps = 8/187 (4%)
Frame = +1
Query: 364 EGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRK 423
EG QDTLY HSLRQFY EC GT+DFIFGN V QNC +Y R+P+ Q ITAQGR
Sbjct: 1 EGYQDTLYVHSLRQFYSECXXXGTVDFIFGNAKVVFQNCNMYPRLPMSGQFNAITAQGRT 180
Query: 424 SPHQSTGFTIQDSYVLASQP--------TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEW 475
P+Q TG +I +S + A+ TYLGRPWKEYSRTVY+ T+M S++ +GW EW
Sbjct: 181 DPNQDTGISIHNSTIRAADDLASSNGVATYLGRPWKEYSRTVYMQTFMDSVIHAKGWREW 360
Query: 476 LGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTG 535
G+FAL TL+Y EY N GPGS RV WPGYHVI +A+ A FTV FL G WLP+TG
Sbjct: 361 DGDFALSTLYYAEYSNSGPGSGTDNRVTWPGYHVI-NATDASNFTVSNFLLGDDWLPQTG 537
Query: 536 VKFTAGL 542
V +T L
Sbjct: 538 VSYTNNL 558
>TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (Fragment) ,
partial (49%)
Length = 1026
Score = 214 bits (546), Expect = 6e-56
Identities = 109/207 (52%), Positives = 133/207 (63%), Gaps = 9/207 (4%)
Frame = +3
Query: 345 QAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKI 404
QAVALRV D SAFF C I QDTLY H+ RQF+ +C I GT+DFIFGN A V Q+C I
Sbjct: 6 QAVALRVGGDLSAFFNCDILAFQDTLYVHNNRQFFEKCLIAGTVDFIFGNSAVVFQDCDI 185
Query: 405 YTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQP---------TYLGRPWKEYSR 455
+ R+P QK +TAQGR P+Q+TG IQ + A+ TYLGRPWKEYSR
Sbjct: 186 HARLPSSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATNDLESVKKNFKTYLGRPWKEYSR 365
Query: 456 TVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASA 515
TV + + +S ++ P GW EW GNFAL TL Y EY+N GPG+ + RV W GY VI DA+
Sbjct: 366 TVIMQSSISDVIDPIGWHEWSGNFALSTLVYREYQNTGPGAGTSNRVTWKGYKVITDAAE 545
Query: 516 AGYFTVQRFLNGGSWLPRTGVKFTAGL 542
A +T F+ G SWL TG F+ GL
Sbjct: 546 ARDYTPGSFIGGSSWLGSTGFPFSLGL 626
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,179,290
Number of Sequences: 63676
Number of extensions: 310131
Number of successful extensions: 1559
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 1466
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1470
length of query: 544
length of database: 12,639,632
effective HSP length: 102
effective length of query: 442
effective length of database: 6,144,680
effective search space: 2715948560
effective search space used: 2715948560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144513.13


