

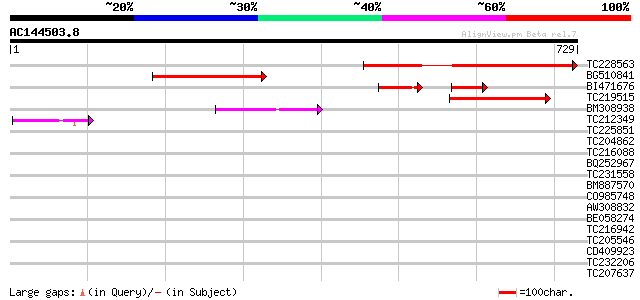
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.8 - phase: 0
(729 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC228563 similar to UP|Q9LTB0 (Q9LTB0) Gb|AAD32890.1 (AT5g49830/... 433 e-121
BG510841 similar to GP|18700079|gb AT5g49830/K21G20_4 {Arabidops... 282 4e-76
BI471676 similar to GP|18700079|gb| AT5g49830/K21G20_4 {Arabidop... 94 9e-41
TC219515 similar to UP|Q9SY60 (Q9SY60) F14N23.6, partial (15%) 129 6e-30
BM308938 weakly similar to GP|27817901|db OJ1092_A07.21 {Oryza s... 82 1e-15
TC212349 similar to UP|Q940L2 (Q940L2) At1g10180/F14N23_6, parti... 44 2e-04
TC225851 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Su... 32 0.77
TC204862 similar to GB|AAM61695.1|21537354|AY085142 hydroxyproli... 31 2.2
TC216088 similar to UP|Q8GYW7 (Q8GYW7) Acyl-CoA thioesterase (At... 30 3.8
BQ252967 30 3.8
TC231558 weakly similar to UP|Q8LQ09 (Q8LQ09) OJ1294_F06.10 prot... 30 3.8
BM887570 30 5.0
CO985748 30 5.0
AW308832 29 6.5
BE058274 similar to GP|20340247|gb| galactinol synthase-like pro... 29 6.5
TC216942 similar to UP|O01721 (O01721) Myosin-like protein (Frag... 29 6.5
TC205546 similar to UP|Q9M621 (Q9M621) ReMembR-H2 protein JR702 ... 29 6.5
CD409923 29 8.5
TC232206 similar to UP|Q8LQP4 (Q8LQP4) OJ1117_G01.16 protein, pa... 29 8.5
TC207637 similar to UP|Q6SJR0 (Q6SJR0) TATA-binding protein asso... 29 8.5
>TC228563 similar to UP|Q9LTB0 (Q9LTB0) Gb|AAD32890.1 (AT5g49830/K21G20_4),
partial (30%)
Length = 1000
Score = 433 bits (1113), Expect = e-121
Identities = 218/275 (79%), Positives = 234/275 (84%)
Frame = +1
Query: 455 DNRRRTTERQNRHPEQREWRRRLVGSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISM 514
DNRRRT+ERQNRHPEQREWRRRLVGSVDRLKD+FCRQHAL LIFTE+GDSHLTADMYI+M
Sbjct: 1 DNRRRTSERQNRHPEQREWRRRLVGSVDRLKDTFCRQHALDLIFTEEGDSHLTADMYINM 180
Query: 515 ERNADEVEWIPSLIFQSVVSWLLIFKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLN 574
+ NA+EVEWIPSLIFQ ELF+KLN
Sbjct: 181 DGNAEEVEWIPSLIFQ-------------------------------------ELFVKLN 249
Query: 575 RMANIAADMFVGRERFATLLLMRLTETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLD 634
RMANIAADMFVGRERFATLLLMRLTETV+LW+SEDQSFWDDIEEGPRPLGPLGLQQFYLD
Sbjct: 250 RMANIAADMFVGRERFATLLLMRLTETVVLWLSEDQSFWDDIEEGPRPLGPLGLQQFYLD 429
Query: 635 MKFVVCFASNGRYLSRNLQRIVNEIIRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMER 694
MKFVVCFAS+GRYLSRNLQRIVNEII KAM+AFSATGMDPY +LPEDEWFN+ICQDAMER
Sbjct: 430 MKFVVCFASHGRYLSRNLQRIVNEIITKAMAAFSATGMDPYRELPEDEWFNDICQDAMER 609
Query: 695 LSGKPKEINGERELSSPTASVSAQSISSVRSHNSS 729
LSGKPKEINGER+L+SPTASVSAQSISSVRSH+SS
Sbjct: 610 LSGKPKEINGERDLNSPTASVSAQSISSVRSHSSS 714
>BG510841 similar to GP|18700079|gb AT5g49830/K21G20_4 {Arabidopsis
thaliana}, partial (19%)
Length = 442
Score = 282 bits (721), Expect = 4e-76
Identities = 144/147 (97%), Positives = 147/147 (99%)
Frame = +1
Query: 184 YQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWATKQT 243
YQYNMQSLRPS+TSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAY+SELVMWATKQT
Sbjct: 1 YQYNMQSLRPSSTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYTSELVMWATKQT 180
Query: 244 EAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQA 303
EAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQA
Sbjct: 181 EAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQA 360
Query: 304 LDANLKRIQESTAAMAAADDWVLTYPP 330
LDANLKRIQESTAA+AAADDWVLTYPP
Sbjct: 361 LDANLKRIQESTAALAAADDWVLTYPP 441
>BI471676 similar to GP|18700079|gb| AT5g49830/K21G20_4 {Arabidopsis
thaliana}, partial (12%)
Length = 421
Score = 94.0 bits (232), Expect(2) = 9e-41
Identities = 44/47 (93%), Positives = 47/47 (99%)
Frame = +2
Query: 568 ELFIKLNRMANIAADMFVGRERFATLLLMRLTETVILWISEDQSFWD 614
ELF+KLNRMANIAADMFVGRERFATLLLMRLTETV+LW+SEDQSFWD
Sbjct: 281 ELFVKLNRMANIAADMFVGRERFATLLLMRLTETVMLWLSEDQSFWD 421
Score = 92.4 bits (228), Expect(2) = 9e-41
Identities = 45/56 (80%), Positives = 50/56 (88%)
Frame = +1
Query: 475 RRLVGSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQ 530
+RLV SVDRLKD+FCRQHAL LIFTE+GDSHLTADMYI+M+ NA EVEW PS IFQ
Sbjct: 1 KRLVSSVDRLKDTFCRQHALDLIFTEEGDSHLTADMYINMDGNA-EVEWTPSSIFQ 165
>TC219515 similar to UP|Q9SY60 (Q9SY60) F14N23.6, partial (15%)
Length = 829
Score = 129 bits (323), Expect = 6e-30
Identities = 60/130 (46%), Positives = 87/130 (66%)
Frame = +1
Query: 566 FEELFIKLNRMANIAADMFVGRERFATLLLMRLTETVILWISEDQSFWDDIEEGPRPLGP 625
F+ LF KL ++A +A D+ +G+E+ +LL RLTET+++W+S++Q FW +E+ PL P
Sbjct: 109 FQALFAKLQQLATVAGDVLLGKEKIQKMLLARLTETLVMWLSDEQEFWGALEDNSAPLKP 288
Query: 626 LGLQQFYLDMKFVVCFASNGRYLSRNLQRIVNEIIRKAMSAFSATGMDPYSDLPEDEWFN 685
LGLQQ LDM F V A Y SR++ +I + I +A+ FSA G+DP S LPEDEWF
Sbjct: 289 LGLQQLILDMHFTVEIARFAGYPSRHIHQIASAITARAIRTFSARGIDPQSALPEDEWFV 468
Query: 686 EICQDAMERL 695
E + A+ +L
Sbjct: 469 ETAKSAINKL 498
>BM308938 weakly similar to GP|27817901|db OJ1092_A07.21 {Oryza sativa
(japonica cultivar-group)}, partial (14%)
Length = 428
Score = 81.6 bits (200), Expect = 1e-15
Identities = 44/138 (31%), Positives = 79/138 (56%)
Frame = +1
Query: 265 RAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQALDANLKRIQESTAAMAAADDW 324
RAA+ +Q +L +CS+LE++GL L +LL L RPSVE+ L++N +R + MA
Sbjct: 1 RAASISIQASLNYCSILESQGLKLSKLLLVLLRPSVEEVLESNFRRARRVVLDMAECCPL 180
Query: 325 VLTYPPNVNRQTGSTTAFQLKLTSSAHRFNLMVQDFFEDVGPLLSMQLGGQALEGLFQVF 384
+ +++ S+++ L S RF +V++ E + P +S+ GG L + Q+F
Sbjct: 181 SPQFASSLSAIASSSSSM---LVESGMRFMHIVEEILEQLTPTVSLHFGGNVLNRILQLF 351
Query: 385 NSYVNMLIKALPESMEEE 402
+ Y++ L +ALP +++
Sbjct: 352 DKYMDALTRALPGPSDDD 405
>TC212349 similar to UP|Q940L2 (Q940L2) At1g10180/F14N23_6, partial (18%)
Length = 521
Score = 44.3 bits (103), Expect = 2e-04
Identities = 33/114 (28%), Positives = 59/114 (50%), Gaps = 10/114 (8%)
Frame = +1
Query: 4 SVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV--HIDSLSISDSDGFS 61
++++ + AF+R S+E +++ EL ++ +S Q L+ L GV ++ + S +D
Sbjct: 184 NMHSKFLAFLRISEEAVEVKHELIELQKHISAQGILVQDLMTGVCRELEEWNQSSNDVAE 363
Query: 62 VNGALDSEHKEISDLDKWLVE--------FPDLLDVLLAERRVEEALAALDEGE 107
+ + E+ +L + L F + +DVLLAE + EEAL ALD E
Sbjct: 364 IQ-----QEPELPELLEPLPNERNDQXXLFLETIDVLLAEHKFEEALEALDAEE 510
>TC225851 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Sucrose-UDP
glucosyltransferase 2) , partial (19%)
Length = 641
Score = 32.3 bits (72), Expect = 0.77
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +2
Query: 608 EDQSFWDDIEEGPRPLGPLGLQQFY---LDMKFVVCFASNGRYLSRNLQRIVNEIIRKA 663
ED S W I +G GLQ+ Y + + + CF+ R LS+N Q V ++ RKA
Sbjct: 230 EDPSHWKKISDG-------GLQRIYER*VILLKLCCFSCETRMLSQNAQVHVEDLFRKA 385
>TC204862 similar to GB|AAM61695.1|21537354|AY085142 hydroxyproline-rich
glycoprotein-like protein {Arabidopsis thaliana;} ,
partial (15%)
Length = 834
Score = 30.8 bits (68), Expect = 2.2
Identities = 34/105 (32%), Positives = 46/105 (43%), Gaps = 6/105 (5%)
Frame = -3
Query: 93 ERRVEEALAALDEGERVVSEAKEMKSLNPSLLLSLQSSITERRQKLADQLAE--AACQPS 150
ER VE AA++E E V S E +L + + E + + + L E AA
Sbjct: 337 ERGVEGGFAAIEEAEGVGSSVVEGANLAFEGFGFVVEVLLEGGEVVGELLPEEGAAGHEG 158
Query: 151 TRGAEL-RASVSALKKL---GDGPHAHSLLLNAHLQRYQYNMQSL 191
+ EL A +S L +L GDGPH L QR Y +SL
Sbjct: 157 SLALELVEARLSQLAQLAWIGDGPHRRELNSKTGEQRKPYLARSL 23
>TC216088 similar to UP|Q8GYW7 (Q8GYW7) Acyl-CoA thioesterase (At1g01710),
partial (87%)
Length = 1767
Score = 30.0 bits (66), Expect = 3.8
Identities = 10/28 (35%), Positives = 23/28 (81%), Gaps = 1/28 (3%)
Frame = -2
Query: 519 DEVEWIPSL-IFQSVVSWLLIFKNYRIR 545
DEV+ +P+ + +++SWL++F+N+++R
Sbjct: 200 DEVKTVPTFSLTNNILSWLIVFENHKLR 117
>BQ252967
Length = 413
Score = 30.0 bits (66), Expect = 3.8
Identities = 17/45 (37%), Positives = 26/45 (57%)
Frame = +1
Query: 531 SVVSWLLIFKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNR 575
+V+ WLL F +R R DDL + L++EC+V F + +L R
Sbjct: 55 AVLGWLLKFGGFRERLLADDLFL-AKLLIECVVIIFTKAAAELER 186
>TC231558 weakly similar to UP|Q8LQ09 (Q8LQ09) OJ1294_F06.10 protein, partial
(14%)
Length = 720
Score = 30.0 bits (66), Expect = 3.8
Identities = 17/45 (37%), Positives = 26/45 (57%)
Frame = +3
Query: 531 SVVSWLLIFKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNR 575
+V+ WLL F +R R DDL + L++EC+V F + +L R
Sbjct: 462 AVLGWLLKFGGFRERLLADDLFL-AKLLIECVVIIFTKAAAELER 593
>BM887570
Length = 423
Score = 29.6 bits (65), Expect = 5.0
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 5/68 (7%)
Frame = +2
Query: 436 LLPRAAMKLSSLNQDPYKDD----NRRRTTERQNRHPEQREWRRRLVGSVDR-LKDSFCR 490
LLP +++K +NQ P ++D N E N P ++ + RL R L++SF +
Sbjct: 224 LLPSSSVKELDINQVPLEEDWMASNMEDEEESSNGEPPRK--KLRLTKEQSRLLEESFRQ 397
Query: 491 QHALSLIF 498
H L+ IF
Sbjct: 398 NHTLNPIF 421
>CO985748
Length = 787
Score = 29.6 bits (65), Expect = 5.0
Identities = 21/54 (38%), Positives = 24/54 (43%), Gaps = 2/54 (3%)
Frame = -3
Query: 64 GALDSEHKEISDLDKWLVEFPDLLDVLLAERRV--EEALAALDEGERVVSEAKE 115
GA + KE SDLD W F LD +L + V E A G V E KE
Sbjct: 485 GANNPVMKEFSDLDSWRSHFSMQLDHVLRDNAVLREPAKLGNHTGSHYVREKKE 324
>AW308832
Length = 451
Score = 29.3 bits (64), Expect = 6.5
Identities = 17/41 (41%), Positives = 22/41 (53%)
Frame = +2
Query: 430 SLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQ 470
S+ L P + S + QD KDDN RTT R+NR E+
Sbjct: 62 SIAVINLFPSGDVPESEIQQD--KDDNLWRTTSRENRKLER 178
>BE058274 similar to GP|20340247|gb| galactinol synthase-like protein
{Thellungiella halophila}, partial (20%)
Length = 327
Score = 29.3 bits (64), Expect = 6.5
Identities = 20/59 (33%), Positives = 32/59 (53%), Gaps = 6/59 (10%)
Frame = -2
Query: 521 VEWIPSLIFQSVVSWLLIFKNYRIRQQIDDLSIHIFLI------VECLVHSFEELFIKL 573
VEWI ++ QS++ LI+ + Q +D S+HIFL+ + L H F ++ I L
Sbjct: 317 VEWI--VVVQSLI---LIYIPPLLHQHLDIFSLHIFLLPCVPPRLRSLSHQFNQIKISL 156
>TC216942 similar to UP|O01721 (O01721) Myosin-like protein (Fragment),
partial (12%)
Length = 1562
Score = 29.3 bits (64), Expect = 6.5
Identities = 42/169 (24%), Positives = 61/169 (35%), Gaps = 5/169 (2%)
Frame = +1
Query: 94 RRVEEALAALDEGERVVSEAKEMKSLNPSLLLSLQSSITERRQKLADQLAEAACQPSTRG 153
R +E+ E + AKE + L+ I + RQK +L EA Q
Sbjct: 250 RTLEKERQRAAESRQEYLAAKEEADTQEGRVRQLEEEIRDIRQKYKQELQEALMQREHLQ 429
Query: 154 AELRASVSALKKLGDGPHAHSLLLNAHLQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSA 213
E+ +A +L HS L+ + N + N S + ++L L S
Sbjct: 430 QEIEKEKAARSELEKTLRVHSSPLSDQTPTTKLN-SAFENGNLSRKLSSASSLGSLEESH 606
Query: 214 VAQAASDSLAIFGE-----EPAYSSELVMWATKQTEAFALLVKRHALAS 257
QA+ DS E E + S V T + AL K LAS
Sbjct: 607 FLQASLDSSDSISERRNIGELSMSPYYVKSMTPSSFEAALRQKEGELAS 753
>TC205546 similar to UP|Q9M621 (Q9M621) ReMembR-H2 protein JR702 (Fragment),
partial (43%)
Length = 1515
Score = 29.3 bits (64), Expect = 6.5
Identities = 30/121 (24%), Positives = 51/121 (41%)
Frame = +1
Query: 190 SLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWATKQTEAFALL 249
S+ S S + A+ + + + A AS +L A SS + + T + AL
Sbjct: 265 SMASSVLSSARSSLASSSAIQIARTASMASSALPSARSSLASSSAIQIARTASMASSALH 444
Query: 250 VKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQALDANLK 309
R +LASS+A RAA++ ++ H +A P + R S Q+ ++
Sbjct: 445 SARSSLASSSAIQISRAASQTPSVSRNH-------SIASTPYVQPSLRSSYHQSPSLSIS 603
Query: 310 R 310
R
Sbjct: 604 R 606
>CD409923
Length = 599
Score = 28.9 bits (63), Expect = 8.5
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Frame = -2
Query: 90 LLAERRVEEALAALDEG---ERVVSEAKEMKSLNPSLLLSLQSSITERRQKLADQLAEAA 146
++AER EEA +DEG ER+V+E + L L +++++ E +LA+A
Sbjct: 295 IVAERAREEAEILMDEGKVEERMVTELFRVLKLMEMDLAMVKAAVKEETLGERLELAKAR 116
Query: 147 CQ 148
C+
Sbjct: 115 CR 110
>TC232206 similar to UP|Q8LQP4 (Q8LQP4) OJ1117_G01.16 protein, partial (34%)
Length = 888
Score = 28.9 bits (63), Expect = 8.5
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Frame = +3
Query: 90 LLAERRVEEALAALDEG---ERVVSEAKEMKSLNPSLLLSLQSSITERRQKLADQLAEAA 146
++AER EEA +DEG ER+V+E + L L +++++ E +LA+A
Sbjct: 462 IVAERAREEAEILMDEGKVEERMVTELFRVLKLMEMDLAMVKAAVKEETLGERLELAKAR 641
Query: 147 CQ 148
C+
Sbjct: 642 CR 647
>TC207637 similar to UP|Q6SJR0 (Q6SJR0) TATA-binding protein associated
factor 4b (Fragment), partial (27%)
Length = 1160
Score = 28.9 bits (63), Expect = 8.5
Identities = 20/70 (28%), Positives = 35/70 (49%), Gaps = 3/70 (4%)
Frame = -2
Query: 177 LNAHLQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAV---AQAASDSLAIFGEEPAYSS 233
L+AH + + S+ P + SY ++T QL+F A+ A++A L + E +
Sbjct: 280 LSAHASFPLHTVTSISPHHCSYPSSHTWP*CQLIFFAMELSARSAFPLLLVQPFEKLLTL 101
Query: 234 ELVMWATKQT 243
+ +WA K T
Sbjct: 100 DYPLWALKIT 71
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.132 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,924,209
Number of Sequences: 63676
Number of extensions: 353464
Number of successful extensions: 1719
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1706
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1717
length of query: 729
length of database: 12,639,632
effective HSP length: 104
effective length of query: 625
effective length of database: 6,017,328
effective search space: 3760830000
effective search space used: 3760830000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144503.8


