

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.7 + phase: 0
(284 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
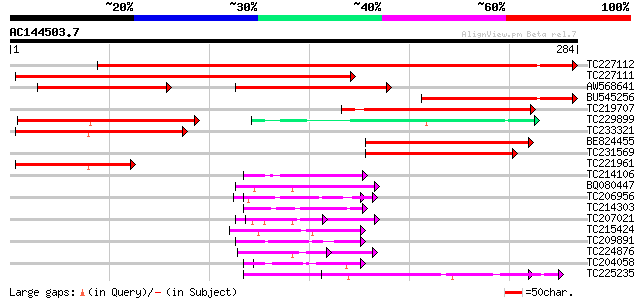
Score E
Sequences producing significant alignments: (bits) Value
TC227112 homologue to UP|Q8L5G3 (Q8L5G3) Proline rich protein 3 ... 455 e-129
TC227111 homologue to UP|Q8L5G3 (Q8L5G3) Proline rich protein 3 ... 311 3e-85
AW568641 homologue to GP|21615411|em proline rich protein 3 {Cic... 159 7e-72
BU545256 122 1e-28
TC219707 weakly similar to GB|AAF55483.4|28381336|AE003720 CG312... 77 1e-14
TC229899 similar to UP|Q9LIC8 (Q9LIC8) Emb|CAA64867.1, partial (... 77 1e-14
TC233321 similar to PIR|T00933|T00933 RNA-binding protein homolo... 75 3e-14
BE824455 72 2e-13
TC231569 similar to UP|Q9LIC8 (Q9LIC8) Emb|CAA64867.1, partial (... 69 2e-12
TC221961 homologue to PIR|T00933|T00933 RNA-binding protein homo... 57 7e-09
TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 51 7e-07
BQ080447 similar to GP|15450467|gb| At1g12810/F13K23_4 {Arabidop... 47 1e-05
TC206956 weakly similar to UP|Q6GUG3 (Q6GUG3) Serine/proline-ric... 46 2e-05
TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, pa... 46 2e-05
TC207021 similar to UP|Q940Z6 (Q940Z6) At1g12810/F13K23_4, parti... 46 2e-05
TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%) 45 3e-05
TC209891 weakly similar to UP|Q9LW52 (Q9LW52) Arabidopsis thalia... 45 3e-05
TC224876 weakly similar to UP|Q39835 (Q39835) Extensin, partial ... 45 3e-05
TC204058 similar to UP|Q39835 (Q39835) Extensin, partial (37%) 45 3e-05
TC225235 similar to GB|AAP37853.1|30725662|BT008494 At1g11650 {A... 45 4e-05
>TC227112 homologue to UP|Q8L5G3 (Q8L5G3) Proline rich protein 3 (Fragment),
partial (78%)
Length = 2000
Score = 455 bits (1171), Expect = e-129
Identities = 216/241 (89%), Positives = 225/241 (92%), Gaps = 1/241 (0%)
Frame = +1
Query: 45 MGFALFNSPHQAIAAKDILQDMLFDPEAKSVLHTEMAKKNLFVKRGIGADAVAFDQSKRL 104
MGFALF++PHQA+ AKDILQDMLFDP+ KSVLHTEMAKKNLFVKRGIGADA AFDQSKRL
Sbjct: 1 MGFALFSAPHQALTAKDILQDMLFDPDTKSVLHTEMAKKNLFVKRGIGADAGAFDQSKRL 180
Query: 105 RTAGDYNHTGYVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSI-A 163
RTAGDY HTGY +PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVA VPMP P I A
Sbjct: 181 RTAGDYTHTGYTSPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAPVPMPTPAPIAA 360
Query: 164 PSSYVPIQNTKDNPPCNTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVCFIE 223
PS+YVP+QNTKDNPPCNTLFIGNLGENINEEEVRGLFSVQPGFKQMKI+RQERHTVCFIE
Sbjct: 361 PSTYVPVQNTKDNPPCNTLFIGNLGENINEEEVRGLFSVQPGFKQMKILRQERHTVCFIE 540
Query: 224 FEDVNSATNVHHNLQGAVIPSSGSIGMRIQYSKNPFGKRKDGTYPMAVPGANGAPVAMTY 283
FEDVNSATNVHHNLQGAVIPSSGSIGMRIQYSKNPFGKRKD P+AVP ANGAP MTY
Sbjct: 541 FEDVNSATNVHHNLQGAVIPSSGSIGMRIQYSKNPFGKRKD-NLPIAVPSANGAPPTMTY 717
Query: 284 Q 284
Q
Sbjct: 718 Q 720
Score = 30.0 bits (66), Expect = 1.2
Identities = 23/83 (27%), Positives = 37/83 (43%)
Frame = +1
Query: 7 TIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDILQDM 66
T+FI L E++ E E++ L PGF+ ++ + + F F + A LQ
Sbjct: 412 TLFIGNLGENINEEEVRGLFSVQPGFKQMKILRQERHTVCFIEFEDVNSATNVHHNLQGA 591
Query: 67 LFDPEAKSVLHTEMAKKNLFVKR 89
+ P + S+ KN F KR
Sbjct: 592 VI-PSSGSIGMRIQYSKNPFGKR 657
>TC227111 homologue to UP|Q8L5G3 (Q8L5G3) Proline rich protein 3 (Fragment),
partial (60%)
Length = 683
Score = 311 bits (796), Expect = 3e-85
Identities = 151/174 (86%), Positives = 160/174 (91%), Gaps = 4/174 (2%)
Frame = +3
Query: 4 QVRTIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDIL 63
+VRTIFITGLPEDVKERELQNL RWLPGFEASQLNFKAEKPMGFALF++PHQA+ AKDIL
Sbjct: 162 EVRTIFITGLPEDVKERELQNLLRWLPGFEASQLNFKAEKPMGFALFSAPHQALTAKDIL 341
Query: 64 QDMLFDPEAKSVLHTEMAKKNLFVKRGIG-ADA-VAFDQSKRLRTAGDYNHTGYVTPSPF 121
QDMLFDP+ KSVLHTEMAKKNLFVKRGIG ADA AFDQSKRLRTAGDY H+GY +PSPF
Sbjct: 342 QDMLFDPDTKSVLHTEMAKKNLFVKRGIGAADAGGAFDQSKRLRTAGDYTHSGYTSPSPF 521
Query: 122 HPPPPPVWGPHGYMA-PPPPPPYDPYAGYPVAQVPMPAPVSI-APSSYVPIQNT 173
HPPPPPVWGPHGYMA PPPPPPYDPYAGYPVA VPMP P I APS+YVP+QNT
Sbjct: 522 HPPPPPVWGPHGYMAPPPPPPPYDPYAGYPVAPVPMPTPAPIPAPSTYVPVQNT 683
Score = 39.7 bits (91), Expect = 0.002
Identities = 43/167 (25%), Positives = 60/167 (35%), Gaps = 1/167 (0%)
Frame = +3
Query: 100 QSKRLRTAGDYNHTGYVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
+ +RL D N P+H W P APPPPPP A P P P
Sbjct: 12 EDRRLGLGFDRNAMAGTGIHPYHQQ----WAPAA-AAPPPPPP-------AAAGGPPPHP 155
Query: 160 VSIAPSSYVPIQNTKDNPPCNTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTV 219
T+FI L E++ E E++ L PGF+ ++ + +
Sbjct: 156 ----------------GEEVRTIFITGLPEDVKERELQNLLRWLPGFEASQLNFKAEKPM 287
Query: 220 CFIEFEDVNSATNVHHNLQGAVI-PSSGSIGMRIQYSKNPFGKRKDG 265
F F + A LQ + P + S+ KN F KR G
Sbjct: 288 GFALFSAPHQALTAKDILQDMLFDPDTKSVLHTEMAKKNLFVKRGIG 428
>AW568641 homologue to GP|21615411|em proline rich protein 3 {Cicer
arietinum}, partial (51%)
Length = 439
Score = 159 bits (403), Expect(2) = 7e-72
Identities = 70/80 (87%), Positives = 73/80 (90%), Gaps = 2/80 (2%)
Frame = +1
Query: 114 GYVTPSPFHPPPPPVWGPHGYMA-PPPPPPYDPYAGYPVAQVPMPAPVSI-APSSYVPIQ 171
GY +PSPFHPPPPPVWGPHGYMA PPPPPPYDPYAGYPVA VPMP P I APS+YVP+Q
Sbjct: 199 GYTSPSPFHPPPPPVWGPHGYMAPPPPPPPYDPYAGYPVAPVPMPTPAPIPAPSTYVPVQ 378
Query: 172 NTKDNPPCNTLFIGNLGENI 191
NTKDNPPCNTLFIGNLGENI
Sbjct: 379 NTKDNPPCNTLFIGNLGENI 438
Score = 129 bits (323), Expect(2) = 7e-72
Identities = 61/67 (91%), Positives = 65/67 (96%)
Frame = +2
Query: 15 EDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDILQDMLFDPEAKS 74
EDVKERELQNL RWLPGFEASQLNFKAEKPMGFALF++PHQA+AAKDILQDMLFDP+ KS
Sbjct: 2 EDVKERELQNLLRWLPGFEASQLNFKAEKPMGFALFSAPHQALAAKDILQDMLFDPDTKS 181
Query: 75 VLHTEMA 81
VLHTEMA
Sbjct: 182VLHTEMA 202
>BU545256
Length = 499
Score = 122 bits (307), Expect = 1e-28
Identities = 62/78 (79%), Positives = 65/78 (82%)
Frame = -1
Query: 207 KQMKIIRQERHTVCFIEFEDVNSATNVHHNLQGAVIPSSGSIGMRIQYSKNPFGKRKDGT 266
KQMKI+ QERHT FIEFEDVNSATNVHHN+QGAVIPSS SIG IQYSKNPFGKRKD
Sbjct: 499 KQMKILSQERHTXXFIEFEDVNSATNVHHNVQGAVIPSSXSIGXXIQYSKNPFGKRKD-N 323
Query: 267 YPMAVPGANGAPVAMTYQ 284
P+AV ANGAP MTYQ
Sbjct: 322 LPVAVHSANGAPXTMTYQ 269
>TC219707 weakly similar to GB|AAF55483.4|28381336|AE003720 CG31243-PE
{Drosophila melanogaster;} , partial (3%)
Length = 673
Score = 76.6 bits (187), Expect = 1e-14
Identities = 40/98 (40%), Positives = 64/98 (64%), Gaps = 1/98 (1%)
Frame = +2
Query: 167 YVPIQNTKDNPPCNTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVCFIEFED 226
Y+P +T PC TLF+ NLG + NE+E+ +FS PGF ++K+ V F++F+D
Sbjct: 83 YIPQNST----PCATLFVANLGPSCNEQELIQVFSRYPGFLKLKMQSTYGAPVAFVDFQD 250
Query: 227 VNSATNVHHNLQGAVIPSSGS-IGMRIQYSKNPFGKRK 263
V S+T+ ++LQG ++ SS S GMR++Y+K+ G R+
Sbjct: 251 VGSSTDALNSLQGTILHSSQSGEGMRLEYAKSRMGMRR 364
Score = 27.3 bits (59), Expect = 8.1
Identities = 20/84 (23%), Positives = 36/84 (42%), Gaps = 1/84 (1%)
Frame = +2
Query: 7 TIFITGLPEDVKERELQNLCRWLPGFEASQLNFKAEKPMGFALFNSPHQAIAAKDILQ-D 65
T+F+ L E+EL + PGF ++ P+ F F + A + LQ
Sbjct: 113 TLFVANLGPSCNEQELIQVFSRYPGFLKLKMQSTYGAPVAFVDFQDVGSSTDALNSLQGT 292
Query: 66 MLFDPEAKSVLHTEMAKKNLFVKR 89
+L ++ + E AK + ++R
Sbjct: 293 ILHSSQSGEGMRLEYAKSRMGMRR 364
>TC229899 similar to UP|Q9LIC8 (Q9LIC8) Emb|CAA64867.1, partial (26%)
Length = 736
Score = 76.6 bits (187), Expect = 1e-14
Identities = 40/93 (43%), Positives = 59/93 (63%), Gaps = 2/93 (2%)
Frame = +1
Query: 5 VRTIFITGLPEDVKERELQNLCRWLPGFEASQLNF--KAEKPMGFALFNSPHQAIAAKDI 62
+ T+F++GLP+DVK RE+ NL R PGF++ QL + +A + + FA F + A+AA
Sbjct: 259 INTLFVSGLPDDVKAREIHNLFRRRPGFDSCQLKYTGRANQVVAFATFFNHQSAMAALHA 438
Query: 63 LQDMLFDPEAKSVLHTEMAKKNLFVKRGIGADA 95
L + FDP+ SVLH E+A+ N KR G +A
Sbjct: 439 LNGVKFDPQTGSVLHIELARSNSRRKRKPGGEA 537
Score = 48.5 bits (114), Expect = 3e-06
Identities = 42/148 (28%), Positives = 59/148 (39%), Gaps = 4/148 (2%)
Frame = +1
Query: 122 HPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNPPCNT 181
H PP AP PP+DPY Y Q + NT
Sbjct: 172 HSPPQTT-------APMAQPPFDPYYLY---------------------QQQDEQSNINT 267
Query: 182 LFIGNLGENINEEEVRGLFSVQPGFK--QMKIIRQERHTVCFIEFEDVNSATNVHHNLQG 239
LF+ L +++ E+ LF +PGF Q+K + V F F + SA H L G
Sbjct: 268 LFVSGLPDDVKAREIHNLFRRRPGFDSCQLKYTGRANQVVAFATFFNHQSAMAALHALNG 447
Query: 240 AVI-PSSGSIGMRIQYSK-NPFGKRKDG 265
P +GS+ + I+ ++ N KRK G
Sbjct: 448 VKFDPQTGSV-LHIELARSNSRRKRKPG 528
>TC233321 similar to PIR|T00933|T00933 RNA-binding protein homolog At2g42240
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(53%)
Length = 699
Score = 75.5 bits (184), Expect = 3e-14
Identities = 40/88 (45%), Positives = 55/88 (62%), Gaps = 2/88 (2%)
Frame = +2
Query: 4 QVRTIFITGLPEDVKERELQNLCRWLPGFEASQLN--FKAEKPMGFALFNSPHQAIAAKD 61
+VRT+F+ GLPEDVK RE+ NL R P +E+S L + +P FA+F S AI AK
Sbjct: 83 EVRTLFVAGLPEDVKPREIYNLFREFPDYESSHLRSPSNSSQPFAFAVFASQQSAIMAKH 262
Query: 62 ILQDMLFDPEAKSVLHTEMAKKNLFVKR 89
L ++FD E S+L+ ++AK N KR
Sbjct: 263 ALNGLVFDLEKGSILYIDLAKSNSRSKR 346
>BE824455
Length = 596
Score = 72.4 bits (176), Expect = 2e-13
Identities = 34/84 (40%), Positives = 52/84 (61%)
Frame = -1
Query: 179 CNTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVCFIEFEDVNSATNVHHNLQ 238
C+TLFI NLG N E+E++ FS GF +K+ + V F++FE+ + A V LQ
Sbjct: 470 CSTLFIANLGPNCTEDELKQAFSAYTGFNMVKMRSRGGMPVAFVDFEETHQAAKVMEELQ 291
Query: 239 GAVIPSSGSIGMRIQYSKNPFGKR 262
G+++PSS GM I+Y+++ KR
Sbjct: 290 GSLLPSSDRGGMHIEYARSKMRKR 219
>TC231569 similar to UP|Q9LIC8 (Q9LIC8) Emb|CAA64867.1, partial (28%)
Length = 605
Score = 69.3 bits (168), Expect = 2e-12
Identities = 33/76 (43%), Positives = 48/76 (62%)
Frame = +1
Query: 179 CNTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVCFIEFEDVNSATNVHHNLQ 238
C+TLFI NLG N E+E++ FSV GF +K+ + V F++FE+ + A V LQ
Sbjct: 7 CSTLFIANLGPNCTEDELKQAFSVYTGFNMVKMRSRGGMPVAFVDFEETDQAAKVVEELQ 186
Query: 239 GAVIPSSGSIGMRIQY 254
G+++PSS GM I+Y
Sbjct: 187 GSLLPSSDRGGMHIEY 234
>TC221961 homologue to PIR|T00933|T00933 RNA-binding protein homolog
At2g42240 - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (29%)
Length = 450
Score = 57.4 bits (137), Expect = 7e-09
Identities = 30/62 (48%), Positives = 39/62 (62%), Gaps = 2/62 (3%)
Frame = +2
Query: 4 QVRTIFITGLPEDVKERELQNLCRWLPGFEASQLN--FKAEKPMGFALFNSPHQAIAAKD 61
+VRT+F+ GLPEDVK RE+ NL R PG+E+S L + +P FA+F S AI A
Sbjct: 257 EVRTLFVAGLPEDVKPREIYNLFREFPGYESSHLRSPSNSSQPFAFAVFASQKSAILAMH 436
Query: 62 IL 63
L
Sbjct: 437 AL 442
Score = 40.4 bits (93), Expect = 0.001
Identities = 38/131 (29%), Positives = 52/131 (39%), Gaps = 6/131 (4%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWG----PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPI 170
Y P P PPPPP+ P G + PP + PY VAQ A + S P
Sbjct: 92 YQQPPP--PPPPPMMAVVPPPPGAVVPPAHHLHPPY----VAQHQQQA---VFGSYGAPQ 244
Query: 171 QNTKDNPPCNTLFIGNLGENINEEEVRGLFSVQPGFK--QMKIIRQERHTVCFIEFEDVN 228
+T + TLF+ L E++ E+ LF PG++ ++ F F
Sbjct: 245 SSTHE---VRTLFVAGLPEDVKPREIYNLFREFPGYESSHLRSPSNSSQPFAFAVFASQK 415
Query: 229 SATNVHHNLQG 239
SA H L G
Sbjct: 416 SAILAMHALNG 448
>TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (22%)
Length = 1296
Score = 50.8 bits (120), Expect = 7e-07
Identities = 29/63 (46%), Positives = 30/63 (47%), Gaps = 1/63 (1%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPV-SIAPSSYVPIQNTKDN 176
P P H PPPP PH +PPPPPP P YP P P PV S P Y P
Sbjct: 208 PPPQHSPPPPP--PH---SPPPPPPSPPPPVYPYLSPPPPPPVHSPPPPVYSPPPPPPSP 372
Query: 177 PPC 179
PPC
Sbjct: 373 PPC 381
Score = 44.3 bits (103), Expect = 6e-05
Identities = 25/62 (40%), Positives = 28/62 (44%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNP 177
P P H PPPPV+ +PPPPPP P P + P P P I P P P
Sbjct: 310 PPPVHSPPPPVY------SPPPPPPSPP----PCIEPPPPPPPCIEPPPPPP-----SPP 444
Query: 178 PC 179
PC
Sbjct: 445 PC 450
Score = 40.0 bits (92), Expect = 0.001
Identities = 26/66 (39%), Positives = 32/66 (48%), Gaps = 4/66 (6%)
Frame = +1
Query: 118 PSPFH--PPPPPVWGP--HGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNT 173
P P H PPPPP P + Y++PPPPPP ++ P P P P S P I+
Sbjct: 232 PPPPHSPPPPPPSPPPPVYPYLSPPPPPPV--HSPPPPVYSPPPPPPSPPPC----IEPP 393
Query: 174 KDNPPC 179
PPC
Sbjct: 394 PPPPPC 411
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/47 (40%), Positives = 21/47 (44%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAP 164
PSP PPPP + +PPPP P Y P P P PV P
Sbjct: 505 PSPSPPPPPVQYNSPPPPSPPPPTPVYHYNSPPPPSFPPPTPVYEGP 645
Score = 38.5 bits (88), Expect = 0.003
Identities = 25/67 (37%), Positives = 27/67 (39%), Gaps = 7/67 (10%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHGYMAPP---PPPPYDPYAGYPVAQVPMPAPV----SIAPSSYVPI 170
P H PPPP P Y PP PPPP Y P P P PV S P S+ P
Sbjct: 445 PCEEHSPPPPSPHPAPYHPPPSPSPPPPPVQYNSPPPPSPPPPTPVYHYNSPPPPSFPPP 624
Query: 171 QNTKDNP 177
+ P
Sbjct: 625 TPVYEGP 645
Score = 38.5 bits (88), Expect = 0.003
Identities = 26/74 (35%), Positives = 32/74 (43%), Gaps = 12/74 (16%)
Frame = +1
Query: 117 TPSPFHP----PPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAP-------- 164
+P P P PPPPV+ P PPPP P P Y V P P+P +P
Sbjct: 1 SPPPPSPPPPSPPPPVYSP----PPPPPSPPPPSPTYCVRSPPPPSPPPPSPPPPPPPVF 168
Query: 165 SSYVPIQNTKDNPP 178
S P+Q +PP
Sbjct: 169 SPPPPVQYYYSSPP 210
Score = 35.8 bits (81), Expect = 0.023
Identities = 23/65 (35%), Positives = 27/65 (41%), Gaps = 3/65 (4%)
Frame = +1
Query: 117 TPSPFHPPPPPVWGPHGYMAPPPPP---PYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNT 173
+P P P PPP P PPPPP P P P + P P S P+ Y P +
Sbjct: 346 SPPPPPPSPPPCIEP----PPPPPPCIEPPPPPPSPPPCEEHSPPPPSPHPAPYHPPPSP 513
Query: 174 KDNPP 178
PP
Sbjct: 514 SPPPP 528
Score = 35.8 bits (81), Expect = 0.023
Identities = 24/67 (35%), Positives = 28/67 (40%), Gaps = 6/67 (8%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHG---YMAPPPPPPYDPYAGYPVAQVPM---PAPVSIAPSSYVPIQ 171
P + PPPPP P Y PPPP P P P+ P PV SS P Q
Sbjct: 40 PPVYSPPPPPPSPPPPSPTYCVRSPPPPSPPPPSPPPPPPPVFSPPPPVQYYYSSPPPPQ 219
Query: 172 NTKDNPP 178
++ PP
Sbjct: 220 HSPPPPP 240
Score = 35.4 bits (80), Expect = 0.030
Identities = 20/62 (32%), Positives = 26/62 (41%), Gaps = 6/62 (9%)
Frame = +1
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSI------APSSYVPIQNTKDN 176
PPPPP P +PPPP P+ P + P P PV +P P+ +
Sbjct: 421 PPPPPSPPPCEEHSPPPPSPHPAPYHPPPSPSPPPPPVQYNSPPPPSPPPPTPVYHYNSP 600
Query: 177 PP 178
PP
Sbjct: 601 PP 606
Score = 34.3 bits (77), Expect = 0.066
Identities = 19/55 (34%), Positives = 28/55 (50%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVP 169
Y +P P PPPP P + PPPP + P PV + P+P + ++ +S P
Sbjct: 538 YNSPPPPSPPPPT---PVYHYNSPPPPSFPPPT--PVYEGPLPPVIGVSYASPPP 687
Score = 32.7 bits (73), Expect = 0.19
Identities = 21/61 (34%), Positives = 25/61 (40%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNP 177
P P PPPP PP PPP + ++ P P PAP PS P + N
Sbjct: 400 PPPCIEPPPP---------PPSPPPCEEHS--PPPPSPHPAPYHPPPSPSPPPPPVQYNS 546
Query: 178 P 178
P
Sbjct: 547 P 549
Score = 31.2 bits (69), Expect = 0.56
Identities = 16/33 (48%), Positives = 18/33 (54%), Gaps = 7/33 (21%)
Frame = +1
Query: 118 PSPFHPPPPPVW-GPH------GYMAPPPPPPY 143
P P PPP PV+ GP Y +PPPPP Y
Sbjct: 601 PPPSFPPPTPVYEGPLPPVIGVSYASPPPPPFY 699
Score = 29.3 bits (64), Expect = 2.1
Identities = 21/61 (34%), Positives = 23/61 (37%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNP 177
PSP PPPPPV+ P PPP Y P P P P + P P
Sbjct: 136 PSP-PPPPPPVFSP--------PPPVQYYYSSPPPPQHSPPP----PPPHSPPPPPPSPP 276
Query: 178 P 178
P
Sbjct: 277 P 279
>BQ080447 similar to GP|15450467|gb| At1g12810/F13K23_4 {Arabidopsis
thaliana}, partial (36%)
Length = 397
Score = 47.0 bits (110), Expect = 1e-05
Identities = 29/76 (38%), Positives = 34/76 (44%), Gaps = 4/76 (5%)
Frame = +3
Query: 114 GYVTPSPF--HPPPPPVWGPHGYMAPPPP--PPYDPYAGYPVAQVPMPAPVSIAPSSYVP 169
GY + P +PPPPP G GY P PP PPYD Y GY P P P +V
Sbjct: 72 GYPSAPPHEGYPPPPPPPGYGGYPPPHPPQRPPYDSYQGYFDNGRPPPPPPPHYHYQHVD 251
Query: 170 IQNTKDNPPCNTLFIG 185
+ D P C + G
Sbjct: 252 HHHLHDEPGCFSFLRG 299
>TC206956 weakly similar to UP|Q6GUG3 (Q6GUG3) Serine/proline-rich repeat
protein, partial (28%)
Length = 788
Score = 46.2 bits (108), Expect = 2e-05
Identities = 25/68 (36%), Positives = 30/68 (43%), Gaps = 2/68 (2%)
Frame = +3
Query: 113 TGYVTP--SPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPI 170
T Y P + +PPPP Y PPPPPP YP+ P S PSSY P
Sbjct: 159 TSYPPPPSATSYPPPPSYHTTGSYSYPPPPPPVASTVSYPLPPTAYPPYSSHPPSSYPPQ 338
Query: 171 QNTKDNPP 178
++ PP
Sbjct: 339 PSSYPPPP 362
Score = 40.8 bits (94), Expect = 7e-04
Identities = 24/67 (35%), Positives = 33/67 (48%)
Frame = +3
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNP 177
P P PPPP + P + PPPPY P AGYP P P P + S++ +TK+
Sbjct: 333 PQPSSYPPPPSYPPPA--SAYPPPPYPPPAGYPPGIYP-PPPY*LGSSTF----STKEKE 491
Query: 178 PCNTLFI 184
+T +
Sbjct: 492 ATSTCLV 512
>TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (18%)
Length = 1014
Score = 46.2 bits (108), Expect = 2e-05
Identities = 25/62 (40%), Positives = 27/62 (43%)
Frame = +2
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNP 177
P P + PPPP P +PPPPPP PY P P P S P Y P P
Sbjct: 38 PPPTYSPPPPPPSPPPPHSPPPPPPVYPYLSPP----PPPPVHSPPPPVYSPPPPPPSPP 205
Query: 178 PC 179
PC
Sbjct: 206 PC 211
Score = 41.6 bits (96), Expect = 4e-04
Identities = 25/62 (40%), Positives = 29/62 (46%)
Frame = +2
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNP 177
P P PPPPP P Y++PPPPPP ++ P P P P S P P P
Sbjct: 80 PPPHSPPPPPPVYP--YLSPPPPPPV--HSPPPPVYSPPPPPPSPPPCIESP-PPPPPPP 244
Query: 178 PC 179
PC
Sbjct: 245 PC 250
Score = 40.0 bits (92), Expect = 0.001
Identities = 27/73 (36%), Positives = 30/73 (40%), Gaps = 12/73 (16%)
Frame = +2
Query: 118 PSPFHPPPPPVWGPHG--------YMAPPPPPPYDPYAGY-PVAQVPMPAPVSIAPSSY- 167
P P H PPPPV+ P +PPPPPP P + P P AP PS
Sbjct: 140 PPPVHSPPPPVYSPPPPPPSPPPCIESPPPPPPPPPCEEHSPPPPSPHSAPYHPPPSPSP 319
Query: 168 --VPIQNTKDNPP 178
PIQ PP
Sbjct: 320 PPPPIQYNSPPPP 358
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/47 (42%), Positives = 22/47 (46%)
Frame = +2
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAP 164
PSP PPPP + +PPPP P Y P P PAPV P
Sbjct: 305 PSPSPPPPPIQYNSPPPPSPPPPTPVYHYNSPPPPSSPPPAPVYEGP 445
Score = 36.2 bits (82), Expect = 0.017
Identities = 20/46 (43%), Positives = 24/46 (51%), Gaps = 1/46 (2%)
Frame = +2
Query: 115 YVTPSPFHPPPPPVWG-PHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
Y++P PPPPPV P +PPPPPP P P + P P P
Sbjct: 122 YLSP----PPPPPVHSPPPPVYSPPPPPPSPP----PCIESPPPPP 235
Score = 34.7 bits (78), Expect = 0.050
Identities = 21/48 (43%), Positives = 21/48 (43%), Gaps = 5/48 (10%)
Frame = +2
Query: 118 PSPFHPPPPPVWGPHG--YMAPP---PPPPYDPYAGYPVAQVPMPAPV 160
P H PPPP PH Y PP PPPP Y P P P PV
Sbjct: 245 PCEEHSPPPP--SPHSAPYHPPPSPSPPPPPIQYNSPPPPSPPPPTPV 382
Score = 34.3 bits (77), Expect = 0.066
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 1/56 (1%)
Frame = +2
Query: 115 YVTPSPFHPPPP-PVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVP 169
Y +P P PPPP PV + Y +PPPP P PV + P+P + ++ +S P
Sbjct: 338 YNSPPPPSPPPPTPV---YHYNSPPPPSSPPP---APVYEGPLPPVIGVSYASPPP 487
Score = 31.2 bits (69), Expect = 0.56
Identities = 15/33 (45%), Positives = 15/33 (45%)
Frame = +2
Query: 137 PPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVP 169
PPPPP Y P A P P P P P S P
Sbjct: 5 PPPPPVYSPPAPPPTYSPPPPPPSPPPPHSPPP 103
Score = 31.2 bits (69), Expect = 0.56
Identities = 16/33 (48%), Positives = 18/33 (54%), Gaps = 7/33 (21%)
Frame = +2
Query: 118 PSPFHPPPPPVW-GPH------GYMAPPPPPPY 143
P P PPP PV+ GP Y +PPPPP Y
Sbjct: 401 PPPSSPPPAPVYEGPLPPVIGVSYASPPPPPFY 499
>TC207021 similar to UP|Q940Z6 (Q940Z6) At1g12810/F13K23_4, partial (51%)
Length = 710
Score = 45.8 bits (107), Expect = 2e-05
Identities = 28/77 (36%), Positives = 34/77 (43%), Gaps = 5/77 (6%)
Frame = +3
Query: 114 GYVTPSP---FHPPPPPVWGPHGYMAPPPP--PPYDPYAGYPVAQVPMPAPVSIAPSSYV 168
GY + P + PPPPP G GY P PP PPYD Y GY P P P +V
Sbjct: 150 GYPSAPPHEGYPPPPPPPHGYGGYPPPQPPQRPPYDSYQGYFDNGHPPPPPPPHYHYQHV 329
Query: 169 PIQNTKDNPPCNTLFIG 185
+ + P C + G
Sbjct: 330 DHHHHHEEPGCFSFLRG 380
Score = 41.6 bits (96), Expect = 4e-04
Identities = 21/43 (48%), Positives = 23/43 (52%), Gaps = 2/43 (4%)
Frame = +3
Query: 119 SPFHPPPP--PVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
SP+ PP P P PH PPPPPP+ Y GYP Q P P
Sbjct: 126 SPYPPPQPGYPSAPPHEGYPPPPPPPHG-YGGYPPPQPPQRPP 251
>TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%)
Length = 1030
Score = 45.4 bits (106), Expect = 3e-05
Identities = 27/78 (34%), Positives = 33/78 (41%), Gaps = 10/78 (12%)
Frame = +3
Query: 111 NHTGYVTPSPFHP------PPPPVWGPHGYMAPPPPPPYDPYAGYP----VAQVPMPAPV 160
+H Y P+P HP PPPP Y PPPPPP Y YP + P P P
Sbjct: 87 HHVPYAPPNPHHPHYPYPPPPPPPPPEASYQPPPPPPPAPMY--YPSNNQYSNQPPPPPP 260
Query: 161 SIAPSSYVPIQNTKDNPP 178
++P P + PP
Sbjct: 261 PLSPPPPPPPVSPPPPPP 314
Score = 38.9 bits (89), Expect = 0.003
Identities = 23/65 (35%), Positives = 26/65 (39%), Gaps = 15/65 (23%)
Frame = +3
Query: 112 HTGYVTPSP-------FHPPPPPVWGPHGYMA--------PPPPPPYDPYAGYPVAQVPM 156
H Y P P + PPPPP P Y + PPPPPP P P P
Sbjct: 126 HYPYPPPPPPPPPEASYQPPPPPPPAPMYYPSNNQYSNQPPPPPPPLSPPPPPPPVSPPP 305
Query: 157 PAPVS 161
P PV+
Sbjct: 306 PPPVT 320
Score = 34.7 bits (78), Expect = 0.050
Identities = 23/60 (38%), Positives = 27/60 (44%), Gaps = 2/60 (3%)
Frame = +3
Query: 115 YVTPSPFHPPPPPVWGPHG--YMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQN 172
Y P P PPP P++ P Y PPPPP P++ P P PVS P V N
Sbjct: 174 YQPPPP--PPPAPMYYPSNNQYSNQPPPPP------PPLSPPPPPPPVSPPPPPPVTQNN 329
Score = 29.3 bits (64), Expect = 2.1
Identities = 16/44 (36%), Positives = 17/44 (38%)
Frame = +3
Query: 110 YNHTGYVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQ 153
Y + P PPPP PPPPPP P PV Q
Sbjct: 213 YPSNNQYSNQPPPPPPP-------LSPPPPPPPVSPPPPPPVTQ 323
>TC209891 weakly similar to UP|Q9LW52 (Q9LW52) Arabidopsis thaliana genomic
DNA, chromosome 3, P1 clone: MLM24, partial (10%)
Length = 646
Score = 45.4 bits (106), Expect = 3e-05
Identities = 27/65 (41%), Positives = 31/65 (47%)
Frame = -3
Query: 114 GYVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNT 173
G P P PPPP+ P G PPPPPP P YP A PMP P +P N+
Sbjct: 338 GLKNPRPGRNPPPPM-PPPGKNPPPPPPPMPPPGKYP-APPPMP-----PPGKKLPPPNS 180
Query: 174 KDNPP 178
+PP
Sbjct: 179 LASPP 165
>TC224876 weakly similar to UP|Q39835 (Q39835) Extensin, partial (66%)
Length = 1340
Score = 45.4 bits (106), Expect = 3e-05
Identities = 22/49 (44%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPP--PPYDPYAGYPVAQVPMPAPVS 161
Y P +H PPPP P+ Y +PPPP PPY P +PV P P P +
Sbjct: 514 YKKPYYYHSPPPPPKKPYKYPSPPPPVHPPYTP---HPVYHSPPPPPTN 651
Score = 42.4 bits (98), Expect = 2e-04
Identities = 24/70 (34%), Positives = 31/70 (44%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
Y SP P PPP + Y PPPPP PY + P P P P + P+ ++
Sbjct: 472 YYYHSPPPPSPPPPYKKPYYYHSPPPPPKKPY------KYPSPPPPVHPPYTPHPVYHSP 633
Query: 175 DNPPCNTLFI 184
PP N +I
Sbjct: 634 PPPPTNKPYI 663
Score = 39.7 bits (91), Expect = 0.002
Identities = 19/45 (42%), Positives = 22/45 (48%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
Y SP P PPP P+ Y +PPPP P P Y P P+P
Sbjct: 370 YYYHSPPPPSPPPPKKPYYYHSPPPPSPSPPKKPYYYHSPPPPSP 504
Score = 39.7 bits (91), Expect = 0.002
Identities = 17/37 (45%), Positives = 20/37 (53%)
Frame = +1
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
PPPPP P+ Y +PPPP P P Y P P+P
Sbjct: 343 PPPPPKKKPYYYHSPPPPSPPPPKKPYYYHSPPPPSP 453
Score = 37.0 bits (84), Expect = 0.010
Identities = 21/49 (42%), Positives = 26/49 (52%), Gaps = 1/49 (2%)
Frame = +3
Query: 112 HTGYVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYP-VAQVPMPAP 159
H YV PSP PPP P+ Y +PPPP + P+ P V P P+P
Sbjct: 927 HPPYVYPSP----PPPPKKPYKYPSPPPPVHHYPHPHPPFVHYSPPPSP 1061
Score = 36.6 bits (83), Expect = 0.013
Identities = 16/40 (40%), Positives = 20/40 (50%)
Frame = +1
Query: 120 PFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
P+H P PP + Y +PPPP P P Y P P+P
Sbjct: 187 PYHYPSPPPPPKYYYHSPPPPSPSPPKKPYYYHSPPPPSP 306
Score = 36.2 bits (82), Expect = 0.017
Identities = 20/54 (37%), Positives = 24/54 (44%), Gaps = 9/54 (16%)
Frame = +1
Query: 115 YVTPSP-----FHPPPPPVWGP----HGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
Y +P P +H PPPP P + Y +PPPP P P Y P P P
Sbjct: 196 YPSPPPPPKYYYHSPPPPSPSPPKKPYYYHSPPPPSPTPPKKPYYYHSPPPPPP 357
Score = 36.2 bits (82), Expect = 0.017
Identities = 21/48 (43%), Positives = 27/48 (55%), Gaps = 3/48 (6%)
Frame = +1
Query: 115 YVTPSPFHPPP---PPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
Y+ SP PPP PPV P+ Y +PPPPP Y ++ P P P+P
Sbjct: 136 YLYSSP--PPPKKYPPVSPPYHYPSPPPPPKYYYHSPPP----PSPSP 261
Score = 36.2 bits (82), Expect = 0.017
Identities = 17/37 (45%), Positives = 20/37 (53%)
Frame = +3
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
PPPPP P+ Y +PPPP PY +P P P P
Sbjct: 789 PPPPPKKKPYYYHSPPPPHPYPHPHPHPYPH-PHPHP 896
Score = 35.0 bits (79), Expect = 0.039
Identities = 16/29 (55%), Positives = 18/29 (61%)
Frame = +3
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPY 143
Y PSP PPPPP P+ Y +PPPP Y
Sbjct: 1071 YKYPSP--PPPPPTNKPYIYASPPPPYHY 1151
Score = 35.0 bits (79), Expect = 0.039
Identities = 18/45 (40%), Positives = 20/45 (44%)
Frame = +1
Query: 115 YVTPSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAP 159
Y SP P P P P+ Y +PPPPPP Y P P P
Sbjct: 274 YYYHSPPPPSPTPPKKPYYYHSPPPPPPKKKPYYYHSPPPPSPPP 408
Score = 34.3 bits (77), Expect = 0.066
Identities = 15/29 (51%), Positives = 18/29 (61%), Gaps = 2/29 (6%)
Frame = +1
Query: 117 TPSPFH--PPPPPVWGPHGYMAPPPPPPY 143
TP P + PPPPP P+ Y +PPPP Y
Sbjct: 607 TPHPVYHSPPPPPTNKPYIYASPPPPYHY 693
Score = 33.9 bits (76), Expect = 0.086
Identities = 29/75 (38%), Positives = 31/75 (40%), Gaps = 11/75 (14%)
Frame = +3
Query: 115 YVTPSPF-HPPPPPVWGPHG-YMAP-PPPPPYDPY----AGYPVAQVPMPAP----VSIA 163
Y P P HP P P PH Y+ P PPPPP PY PV P P P S
Sbjct: 873 YPHPHPHPHPHPHPHPHPHPPYVYPSPPPPPKKPYKYPSPPPPVHHYPHPHPPFVHYSPP 1052
Query: 164 PSSYVPIQNTKDNPP 178
PS P + PP
Sbjct: 1053PSPKKPYKYPSPPPP 1097
Score = 32.3 bits (72), Expect = 0.25
Identities = 14/30 (46%), Positives = 18/30 (59%), Gaps = 1/30 (3%)
Frame = +3
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYD-PY 146
P + PPP P+ Y +PPPPPP + PY
Sbjct: 1029 PFVHYSPPPSPKKPYKYPSPPPPPPTNKPY 1118
Score = 30.0 bits (66), Expect = 1.2
Identities = 19/52 (36%), Positives = 24/52 (45%)
Frame = +3
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVP 169
P +H PPPP PH + P P P P+ +P P P P + PS P
Sbjct: 813 PYYYHSPPPPHPYPHPHPHPYPHPHPHPHP-HPHPH-PHPHPPYVYPSPPPP 962
Score = 28.9 bits (63), Expect = 2.8
Identities = 20/64 (31%), Positives = 23/64 (35%), Gaps = 3/64 (4%)
Frame = +1
Query: 118 PSPFHPPPPPVWGPHG---YMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTK 174
P +H PPPP P Y PPPPP Y + P P P Y
Sbjct: 271 PYYYHSPPPPSPTPPKKPYYYHSPPPPPPKKKPYYYHSPPPPSPPPPKKPYYYHSPPPPS 450
Query: 175 DNPP 178
+PP
Sbjct: 451 PSPP 462
>TC204058 similar to UP|Q39835 (Q39835) Extensin, partial (37%)
Length = 852
Score = 45.4 bits (106), Expect = 3e-05
Identities = 24/60 (40%), Positives = 28/60 (46%), Gaps = 4/60 (6%)
Frame = +3
Query: 123 PPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSY----VPIQNTKDNPP 178
PPPPP P+ Y +PPPPP Y + P + P P P P Y PI K PP
Sbjct: 189 PPPPPPEKPYKYPSPPPPPVYKYKSPPPPYKYPSPPPPPKKPYKYPSPPPPIYKYKSPPP 368
Score = 41.2 bits (95), Expect = 5e-04
Identities = 25/61 (40%), Positives = 29/61 (46%)
Frame = +3
Query: 118 PSPFHPPPPPVWGPHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQNTKDNP 177
P + PPPPV + Y +PPPPPP PY YP P P PV S P + P
Sbjct: 141 PYKYKSPPPPV---YKYKSPPPPPPEKPYK-YP---SPPPPPVYKYKSPPPPYKYPSPPP 299
Query: 178 P 178
P
Sbjct: 300 P 302
Score = 37.7 bits (86), Expect = 0.006
Identities = 26/74 (35%), Positives = 31/74 (41%), Gaps = 5/74 (6%)
Frame = +3
Query: 110 YNHTGYVTPSPFHPPPPPVWGPHGYMAPPPP-----PPYDPYAGYPVAQVPMPAPVSIAP 164
Y + P + PPPP P+ Y +PPPP P PY YP P P P P
Sbjct: 249 YKYKSPPPPYKYPSPPPPPKKPYKYPSPPPPIYKYKSPPPPYK-YP---SPPPPPYKY-P 413
Query: 165 SSYVPIQNTKDNPP 178
S P+ K PP
Sbjct: 414 SPPPPVYKYKSPPP 455
Score = 37.0 bits (84), Expect = 0.010
Identities = 27/73 (36%), Positives = 32/73 (42%), Gaps = 9/73 (12%)
Frame = +3
Query: 115 YVTPSPFHPPPPPVW------GPHGYMAPPPPP--PYD-PYAGYPVAQVPMPAPVSIAPS 165
Y PSP PPPPV+ P+ Y +PPPPP PY P P+ + P P PS
Sbjct: 216 YKYPSP---PPPPVYKYKSPPPPYKYPSPPPPPKKPYKYPSPPPPIYKYKSPPPPYKYPS 386
Query: 166 SYVPIQNTKDNPP 178
P PP
Sbjct: 387 PPPPPYKYPSPPP 425
Score = 35.8 bits (81), Expect = 0.023
Identities = 21/67 (31%), Positives = 27/67 (39%), Gaps = 6/67 (8%)
Frame = +3
Query: 118 PSPFHPPPPPVWG------PHGYMAPPPPPPYDPYAGYPVAQVPMPAPVSIAPSSYVPIQ 171
P + PPPP++ P+ Y +PPPPP P PV + P P S P
Sbjct: 312 PYKYPSPPPPIYKYKSPPPPYKYPSPPPPPYKYPSPPPPVYKYKSPPPPDYKYKSPPPPY 491
Query: 172 NTKDNPP 178
PP
Sbjct: 492 KYPSPPP 512
Score = 32.7 bits (73), Expect = 0.19
Identities = 20/67 (29%), Positives = 26/67 (37%), Gaps = 9/67 (13%)
Frame = +3
Query: 110 YNHTGYVTPSPFHPPPPPVWGPHGYMAPPPP---------PPYDPYAGYPVAQVPMPAPV 160
Y + P + PPPP P+ Y +PPPP P Y + P + P P P
Sbjct: 345 YKYKSPPPPYKYPSPPPP---PYKYPSPPPPVYKYKSPPPPDYKYKSPPPPYKYPSPPPT 515
Query: 161 SIAPSSY 167
P Y
Sbjct: 516 PKKPYKY 536
Score = 31.2 bits (69), Expect = 0.56
Identities = 13/35 (37%), Positives = 17/35 (48%)
Frame = +3
Query: 109 DYNHTGYVTPSPFHPPPPPVWGPHGYMAPPPPPPY 143
DY + P + PPP P+ Y +PPPP Y
Sbjct: 459 DYKYKSPPPPYKYPSPPPTPKKPYKYSSPPPPYHY 563
>TC225235 similar to GB|AAP37853.1|30725662|BT008494 At1g11650 {Arabidopsis
thaliana;} , partial (64%)
Length = 1154
Score = 45.1 bits (105), Expect = 4e-05
Identities = 42/152 (27%), Positives = 63/152 (40%), Gaps = 7/152 (4%)
Frame = +3
Query: 118 PSPFHPPPPPVWGPHG-YMAPPPPPPYDPYAGYPVA-QVPMPAPVSIAPSSYVPI-QNTK 174
P P PP P Y PPP PY P Q P P APS+ P+ Q
Sbjct: 54 PGPGMAPPTMGQQPQQQYQQQPPPQQQQPYVMMPPQHQAPQPM---WAPSAQPPLPQQPA 224
Query: 175 DNPPCNTLFIGNLGENINEEEVRGLFSVQPGFKQMKIIRQERHTVC----FIEFEDVNSA 230
TL+IG+L ++E + F+ +K+IR ++ + FIEF A
Sbjct: 225 SADEVRTLWIGDLQYWMDENYLYTCFAHTGEVSSVKVIRNKQTSQSEGYGFIEFNSRAGA 404
Query: 231 TNVHHNLQGAVIPSSGSIGMRIQYSKNPFGKR 262
+ GA++P+ G R+ ++ G+R
Sbjct: 405 ERILQTYNGAIMPNGGQ-SFRLNWATFSAGER 497
Score = 43.1 bits (100), Expect = 1e-04
Identities = 36/125 (28%), Positives = 60/125 (47%), Gaps = 4/125 (3%)
Frame = +3
Query: 157 PAPVSIAPSSYV---PIQNTKDNPPCNT-LFIGNLGENINEEEVRGLFSVQPGFKQMKII 212
PA S +SY+ P + +N P NT +F+GNL N+ ++ +R +FS +KI
Sbjct: 768 PATQSQPKASYLNSQPQGSQNENDPNNTTIFVGNLDPNVTDDHLRQVFSQYGELVHVKIP 947
Query: 213 RQERHTVCFIEFEDVNSATNVHHNLQGAVIPSSGSIGMRIQYSKNPFGKRKDGTYPMAVP 272
+R F++F D + A L G ++ G +R+ + ++P K+ P
Sbjct: 948 AGKR--CGFVQFADRSCAEEALRVLNGTLL---GGQNVRLSWGRSPSNKQAQAD-PNQWN 1109
Query: 273 GANGA 277
GA GA
Sbjct: 1110GAAGA 1124
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,987,501
Number of Sequences: 63676
Number of extensions: 375762
Number of successful extensions: 15412
Number of sequences better than 10.0: 940
Number of HSP's better than 10.0 without gapping: 4968
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8863
length of query: 284
length of database: 12,639,632
effective HSP length: 96
effective length of query: 188
effective length of database: 6,526,736
effective search space: 1227026368
effective search space used: 1227026368
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144503.7


