

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.2 + phase: 0
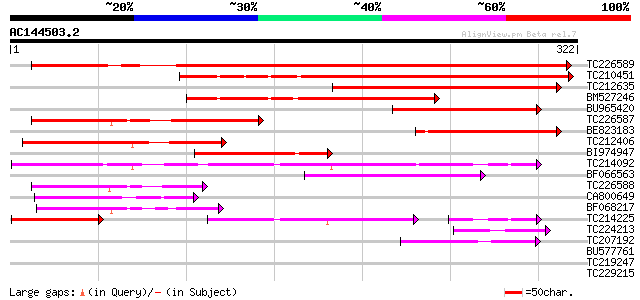
(322 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226589 412 e-115
TC210451 275 3e-74
TC212635 202 2e-52
BM527246 163 1e-40
BU965420 133 1e-31
TC226587 131 4e-31
BE823183 118 4e-27
TC212406 109 1e-24
BI974947 104 6e-23
TC214092 similar to UP|Q9FJY6 (Q9FJY6) Apospory-associated prote... 95 5e-20
BF066563 weakly similar to GP|8777366|dbj| apospory-associated p... 86 2e-17
TC226588 84 7e-17
CA800649 weakly similar to GP|16612258|gb| AT3g01590/F4P13_13 {A... 69 3e-12
BF068217 weakly similar to GP|8777366|dbj| apospory-associated p... 52 3e-07
TC214225 similar to UP|Q9FJY6 (Q9FJY6) Apospory-associated prote... 46 3e-07
TC224213 weakly similar to GB|AAM63374.1|21554299|AY086170 aposp... 49 2e-06
TC207192 44 1e-04
BU577761 40 0.001
TC219247 38 0.007
TC229215 37 0.012
>TC226589
Length = 1426
Score = 412 bits (1058), Expect = e-115
Identities = 200/307 (65%), Positives = 237/307 (77%)
Frame = +2
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCFDM 72
G+NGL+K+ILR+ RG SAEVYLYG VTSWKN+ EELLF+SSK + D
Sbjct: 71 GINGLDKVILRDARGSSAEVYLYGAHVTSWKNDHAEELLFLSSK-------FSNLGPLDS 229
Query: 73 NNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDTK 132
+ F +N+ W +D +PPPF TN+ S+AFVDL+LK SEDD K
Sbjct: 230 HG--------------------FARNRFWTIDDSPPPFLTNTPSKAFVDLILKPSEDDIK 349
Query: 133 NWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLE 192
WPH +EFRLR+ALG GDLM+TSRIRNTN DGK F+FTFA HTY V+DISEVR+EGLE
Sbjct: 350 IWPHSFEFRLRVALGSGGDLMMTSRIRNTNIDGKPFSFTFANHTYFSVSDISEVRVEGLE 529
Query: 193 TLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAV 252
TLDYLD+L+ RERFTEQGDA+TFE E D++YLSTPTKIAI+DHE+KRT +RKDGLPDAV
Sbjct: 530 TLDYLDNLQKRERFTEQGDALTFESEFDRIYLSTPTKIAILDHEKKRTIVLRKDGLPDAV 709
Query: 253 VWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCSGQ 312
VWNPWDKK+K +SD GDDEYK+MLCV+ A +EK ITLKPGEEWKGR E+S VPSSYCSGQ
Sbjct: 710 VWNPWDKKAKAMSDFGDDEYKYMLCVEAAAIEKPITLKPGEEWKGRLELSTVPSSYCSGQ 889
Query: 313 LDPRKVL 319
LDP+KV+
Sbjct: 890 LDPKKVI 910
>TC210451
Length = 921
Score = 275 bits (702), Expect = 3e-74
Identities = 138/224 (61%), Positives = 171/224 (75%)
Frame = +3
Query: 97 KNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTS 156
+N++W LD +P P P N + VDL+LK++E D K P +EFRLRI+L G L+L
Sbjct: 3 RNRMWSLDRDPSPLPPLDNQSS-VDLILKSTEVDLKT-PCSFEFRLRISLN-AGKLILIP 173
Query: 157 RIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFE 216
R+RNT K+F+F+F YL V+DISEVRIEGLETLDY D+L NR RFTEQ DA+TF+
Sbjct: 174 RVRNTAN--KAFSFSFTLCNYLSVSDISEVRIEGLETLDYFDNLLNRSRFTEQADALTFD 347
Query: 217 GEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHML 276
GE+D+VYL +PTKIAIIDHE+KRTF ++K G+PDAVVWNPW KK+K I DLGDD+YK M+
Sbjct: 348 GEMDRVYLHSPTKIAIIDHEKKRTFVLQKKGMPDAVVWNPWGKKAKAIPDLGDDDYKIMM 527
Query: 277 CVQPACVEKAITLKPGEEWKGRQEISAVPSSYCSGQLDPRKVLF 320
CV A ++ I LKP EEW G QE+S V SSYCSGQLDPRKVL+
Sbjct: 528 CVNSAAIDTPILLKPSEEWMGYQELSTVSSSYCSGQLDPRKVLY 659
>TC212635
Length = 837
Score = 202 bits (514), Expect = 2e-52
Identities = 93/130 (71%), Positives = 110/130 (84%)
Frame = +1
Query: 184 SEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEI 243
SE+RIEGLETLDYLD+L +ERFTEQGDAITFE EVD+VYLS+P IA++DHERKRTF I
Sbjct: 133 SEIRIEGLETLDYLDNLFQKERFTEQGDAITFESEVDRVYLSSPNIIAVLDHERKRTFVI 312
Query: 244 RKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISA 303
RKDGLPD VWNPW+KKSK ++D GD+EYKHMLCV A +EK + LKPGEEW GR ++S
Sbjct: 313 RKDGLPDVAVWNPWEKKSKSMADFGDEEYKHMLCVDGAVIEKPVNLKPGEEWTGRLQLSV 492
Query: 304 VPSSYCSGQL 313
VPSS+ S +L
Sbjct: 493 VPSSFFSDRL 522
>BM527246
Length = 422
Score = 163 bits (412), Expect = 1e-40
Identities = 83/144 (57%), Positives = 109/144 (75%)
Frame = +1
Query: 101 WILDPNPPPFPTNSNSRAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRN 160
W LD +P P P + N + VDL++K++ D KN P +EFRLRI+L G L+L R+RN
Sbjct: 1 WSLDRDPSPLPPSDNQSS-VDLIIKSTGVDLKNRPCSFEFRLRISLS-AGKLILIPRVRN 174
Query: 161 TNKDGKSFTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVD 220
T D K+ +FT + YL V+DISEVR+EGLETLDYLD+L NR RFTEQ DAITF+G++D
Sbjct: 175 T--DNKTLSFTLSISNYLSVSDISEVRVEGLETLDYLDNLLNRSRFTEQADAITFDGQMD 348
Query: 221 KVYLSTPTKIAIIDHERKRTFEIR 244
++YL TPTKIA+IDHE+KRTF ++
Sbjct: 349 RLYLRTPTKIAVIDHEKKRTFVLQ 420
>BU965420
Length = 453
Score = 133 bits (334), Expect = 1e-31
Identities = 58/85 (68%), Positives = 70/85 (82%)
Frame = +3
Query: 218 EVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLC 277
+VD+VYLS+P IA++DHERKRTF IRKDGLPD VWNPW+KKSK +SD GD+EYK MLC
Sbjct: 195 QVDRVYLSSPNIIAVLDHERKRTFVIRKDGLPDVAVWNPWEKKSKSMSDFGDEEYKQMLC 374
Query: 278 VQPACVEKAITLKPGEEWKGRQEIS 302
V A +EK + LKPGEEW GR ++S
Sbjct: 375 VDGAVIEKPVNLKPGEEWTGRLQLS 449
>TC226587
Length = 566
Score = 131 bits (330), Expect = 4e-31
Identities = 68/136 (50%), Positives = 90/136 (66%), Gaps = 4/136 (2%)
Frame = +3
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYN 68
G+NGL+K+ILR+ RG SAEVYLYG VTSWKN+ EELLF+SSK P K ++
Sbjct: 195 GINGLDKVILRDARGSSAEVYLYGAHVTSWKNDHAEELLFLSSKAIFKPPKPIRGGIPI- 371
Query: 69 CFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSE 128
CF +N L + F +N+ W +D +PPPFPT+++++AFVDL+LK SE
Sbjct: 372 CFPQFSNLGPLD-----------SHGFARNQFWTIDDSPPPFPTSTSNKAFVDLILKPSE 518
Query: 129 DDTKNWPHRYEFRLRI 144
DD K WPH +EFRLR+
Sbjct: 519 DDIKIWPHSFEFRLRV 566
>BE823183
Length = 589
Score = 118 bits (295), Expect = 4e-27
Identities = 51/83 (61%), Positives = 65/83 (77%)
Frame = -1
Query: 231 AIIDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLK 290
+++DH + F IRKDGLPD VWNPW+KKSK ++D GD+EYKHMLCV A +EK + LK
Sbjct: 556 SVLDHX-EXXFVIRKDGLPDVAVWNPWEKKSKSMADFGDEEYKHMLCVDGAVIEKPVNLK 380
Query: 291 PGEEWKGRQEISAVPSSYCSGQL 313
PGEEW GR ++S VPSS+CS +L
Sbjct: 379 PGEEWTGRLQLSVVPSSFCSDRL 311
>TC212406
Length = 585
Score = 109 bits (273), Expect = 1e-24
Identities = 59/119 (49%), Positives = 75/119 (62%), Gaps = 3/119 (2%)
Frame = +3
Query: 8 VKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQY 67
V+ C GVNGLEKI+LRE+RG S EVYLYGG VTSWKN+ GEELLF+S+K + K +
Sbjct: 261 VELCKGVNGLEKILLRESRGSSTEVYLYGGHVTSWKNDHGEELLFLSNKAIFKTPKAIRG 440
Query: 68 N---CFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLV 123
CF L Q+ F +N+ W +D +PPPFPTN+ S+AFVDL+
Sbjct: 441 GIPLCFPQFGGLGTL-----------DQHGFARNRFWAIDDDPPPFPTNTLSKAFVDLI 584
>BI974947
Length = 402
Score = 104 bits (259), Expect = 6e-23
Identities = 49/79 (62%), Positives = 62/79 (78%), Gaps = 1/79 (1%)
Frame = +1
Query: 106 NPPPFPTNSNS-RAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKD 164
NPPP P N +S ++F+DLVLK+SE+D K WPH +EFRLR++L GDL L SR+RN N
Sbjct: 4 NPPPLPANDSSGKSFIDLVLKSSEEDMKGWPHSFEFRLRVSLTTDGDLTLISRVRNIN-- 177
Query: 165 GKSFTFTFAYHTYLYVTDI 183
GK F+F+FAYHTYL V+DI
Sbjct: 178 GKPFSFSFAYHTYLMVSDI 234
>TC214092 similar to UP|Q9FJY6 (Q9FJY6) Apospory-associated protein C-like
protein, partial (89%)
Length = 1287
Score = 94.7 bits (234), Expect = 5e-20
Identities = 80/308 (25%), Positives = 133/308 (42%), Gaps = 7/308 (2%)
Frame = +1
Query: 2 SRETMYVKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEV 61
S ++ V+ G L K++L G AE+YL+GG +TSWK G++LLFV +P+
Sbjct: 259 STTSLGVRVTEGEGNLPKLVLTSPAGSEAEIYLFGGCITSWKVPSGKDLLFV--RPDAVF 432
Query: 62 KKTNQYN-----CFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNS 116
+ CF Q+ F +N W + + N+
Sbjct: 433 NGNKPISGGVPHCFPQFGP------------GPIQQHGFARNMDWTVVDSE-----NTEG 561
Query: 117 RAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHT 176
V L LK++ W + ++ L L + + N D K+F+F+ A HT
Sbjct: 562 NPVVTLELKDAPYSRAMWDFSFHALFKVTLNAKS---LATELTVKNTDNKAFSFSTALHT 732
Query: 177 YLYVT--DISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIID 234
Y + + S ++G +TL+ KN E+ D +TF G VD VYL +++ +D
Sbjct: 733 YFRASASNASVKGLKGCKTLNKDPDPKNPVEGKEERDVVTFPGFVDCVYLDASSELQ-LD 909
Query: 235 HERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEE 294
+ I+ DAV+WNP+ + YK +CV+ A + ++ L+P +
Sbjct: 910 NGLGDLISIKNTNWSDAVLWNPYLQMEAC--------YKDFVCVENAKI-GSVQLEPEQT 1062
Query: 295 WKGRQEIS 302
W Q +S
Sbjct: 1063WTAVQHLS 1086
>BF066563 weakly similar to GP|8777366|dbj| apospory-associated protein C
{Arabidopsis thaliana}, partial (17%)
Length = 310
Score = 85.9 bits (211), Expect = 2e-17
Identities = 44/103 (42%), Positives = 62/103 (59%)
Frame = +2
Query: 168 FTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTP 227
F+F+ AYHTYL V +I+ V +E L TL YLD L R RFT QGDA TF+ + D +Y++ P
Sbjct: 2 FSFSLAYHTYLSVDNIN*VLLERLNTLVYLDHLYKRHRFT*QGDAHTFQSDCDTIYVTPP 181
Query: 228 TKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDD 270
+ + ++DH RT D + DA+V N D + + DD
Sbjct: 182 SLLGLVDHYNNRTIYSHYDAVSDAIVSNGSDITPIAVGGIIDD 310
>TC226588
Length = 501
Score = 84.3 bits (207), Expect = 7e-17
Identities = 47/104 (45%), Positives = 61/104 (58%), Gaps = 4/104 (3%)
Frame = +2
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSS----KPEKEVKKTNQYN 68
G+NGL+K+ILR+ RG SAEVYLYG VTSWKNE+ EELLF+SS KP K ++
Sbjct: 224 GINGLDKVILRDPRGSSAEVYLYGAHVTSWKNEQAEELLFLSSKAIFKPPKPIRGGIPI- 400
Query: 69 CFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPT 112
CF +N + F +N+ W +D +PPPF T
Sbjct: 401 CFPQFSN-----------LGPLDSHGFARNRFWTIDDSPPPFLT 499
>CA800649 weakly similar to GP|16612258|gb| AT3g01590/F4P13_13 {Arabidopsis
thaliana}, partial (30%)
Length = 436
Score = 68.9 bits (167), Expect = 3e-12
Identities = 40/93 (43%), Positives = 54/93 (58%)
Frame = +2
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCFDMNN 74
+GL +IIL E +G SAEV LYGGQ+ SWKN R EELLF+SSK K+ K +
Sbjct: 182 DGLPRIILTEPKGSSAEVLLYGGQIISWKNHRKEELLFMSSKANKKQHKA-------IRG 340
Query: 75 NKCRLILLLQDLYAEAFQYVFLKNKLWILDPNP 107
+ DL + Q+ F +N++W LD +P
Sbjct: 341 GISACLARFGDL-SSLEQHGFARNRMWSLDRDP 436
>BF068217 weakly similar to GP|8777366|dbj| apospory-associated protein C
{Arabidopsis thaliana}, partial (25%)
Length = 295
Score = 52.4 bits (124), Expect = 3e-07
Identities = 38/110 (34%), Positives = 55/110 (49%), Gaps = 4/110 (3%)
Frame = +2
Query: 16 GLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCFD 71
GL K IL + RG AEV+LYG VTSW NE+ EELLF+S + P ++ C
Sbjct: 2 GLYKDILLDPRGSYAEVFLYGAHVTSWMNEQAEELLFLSGEAMFMPP*PIRGGIPI-C*T 178
Query: 72 MNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVD 121
+ N + L ++ F +N+ W +D P P TN+ + V+
Sbjct: 179 LF*N-------VGPLDSDG----FARNRFWTIDDGPAPLLTNTAGKPMVE 295
>TC214225 similar to UP|Q9FJY6 (Q9FJY6) Apospory-associated protein C-like
protein, partial (86%)
Length = 1334
Score = 46.2 bits (108), Expect = 2e-05
Identities = 22/52 (42%), Positives = 32/52 (61%)
Frame = +3
Query: 2 SRETMYVKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFV 53
S ++ V+ G L K++L G AE+YL+GG +TSWK G++LLFV
Sbjct: 57 SSTSLGVRLTEGEGNLPKLVLTSPAGSEAEIYLFGGFITSWKVPSGKDLLFV 212
Score = 42.7 bits (99), Expect(2) = 3e-07
Identities = 31/122 (25%), Positives = 54/122 (43%), Gaps = 2/122 (1%)
Frame = +2
Query: 113 NSNSRAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTF 172
N+ V L LK+ W + ++ L L + + N D K+F+F+
Sbjct: 563 NT*GNPVVTLELKDGPYSCTVWDFSFHALFKVTLNAKS---LATELTVKNTDNKAFSFST 733
Query: 173 AYHTYLY--VTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKI 230
HTY V++ S ++G +TL+ KN E+ + +TF G VD YL +++
Sbjct: 734 TLHTYFQASVSNASVKGLKGCKTLNKDPDPKNPV*GKEEREVVTFPGFVDCAYLDASSEL 913
Query: 231 AI 232
+
Sbjct: 914 QL 919
Score = 28.9 bits (63), Expect(2) = 3e-07
Identities = 16/53 (30%), Positives = 27/53 (50%)
Frame = +3
Query: 250 DAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEIS 302
DAV+WNP+ + YK +CV+ A + ++ L+P + W Q +S
Sbjct: 966 DAVLWNPYLQMEAC--------YKDFVCVENAKI-GSVQLEPEQTWTAVQHLS 1097
>TC224213 weakly similar to GB|AAM63374.1|21554299|AY086170
apospory-associated protein C {Arabidopsis thaliana;} ,
partial (15%)
Length = 418
Score = 49.3 bits (116), Expect = 2e-06
Identities = 27/55 (49%), Positives = 31/55 (56%)
Frame = +1
Query: 253 VWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
VWNPW+KK K DLGD EYK +EK ITL GE G E+ VPS+
Sbjct: 142 VWNPWEKKYKATVDLGDKEYK--------AIEKPITLNSGE**TGCLELYMVPST 282
>TC207192
Length = 1307
Score = 43.5 bits (101), Expect = 1e-04
Identities = 24/79 (30%), Positives = 36/79 (45%)
Frame = +3
Query: 223 YLSTPTKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPAC 282
Y + P+K ID R F + + G D + +P K D + +C PA
Sbjct: 789 YNTPPSKYETIDQGRGLCFRVIRMGFEDIYLSSPGSLSEKYGKD-------YFICTGPAS 947
Query: 283 VEKAITLKPGEEWKGRQEI 301
+ +T+ PGEEW+G Q I
Sbjct: 948 ILVPVTVNPGEEWRGAQVI 1004
>BU577761
Length = 424
Score = 40.4 bits (93), Expect = 0.001
Identities = 17/20 (85%), Positives = 20/20 (100%)
Frame = +1
Query: 300 EISAVPSSYCSGQLDPRKVL 319
E+SAVPSSYCSGQLDP++VL
Sbjct: 4 ELSAVPSSYCSGQLDPQRVL 63
>TC219247
Length = 1272
Score = 37.7 bits (86), Expect = 0.007
Identities = 26/113 (23%), Positives = 53/113 (46%)
Frame = +1
Query: 189 EGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGL 248
EG ++ + + + N ++ +E+ + VY P +ID R+ + + + G
Sbjct: 808 EGSQSEEMDEEIDNYKQLSEK---------LSHVYTDVPRNFTVIDRGRRNSVSVGRTGF 960
Query: 249 PDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEI 301
+ +++P + +I S +Y + +CV A + K I L P + W+G Q I
Sbjct: 961 DEMYLFSP-GSRVEIYS-----KYSY-ICVGQAAILKPIILSPEDVWRGGQYI 1098
>TC229215
Length = 764
Score = 37.0 bits (84), Expect = 0.012
Identities = 17/22 (77%), Positives = 20/22 (90%)
Frame = +3
Query: 298 RQEISAVPSSYCSGQLDPRKVL 319
R E+SAVPSSY SGQLDP++VL
Sbjct: 3 RLELSAVPSSYRSGQLDPQRVL 68
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,901,739
Number of Sequences: 63676
Number of extensions: 210261
Number of successful extensions: 1210
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 1188
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1196
length of query: 322
length of database: 12,639,632
effective HSP length: 97
effective length of query: 225
effective length of database: 6,463,060
effective search space: 1454188500
effective search space used: 1454188500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144503.2


