

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.1 + phase: 0 /pseudo
(718 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
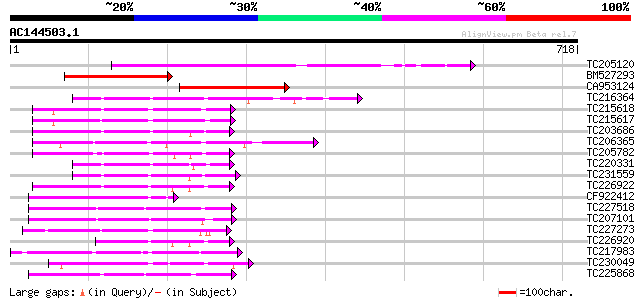
Score E
Sequences producing significant alignments: (bits) Value
TC205120 weakly similar to UP|Q7KSD3 (Q7KSD3) CG7693-PA (Cg7693-... 363 e-100
BM527293 241 6e-64
CA953124 208 6e-54
TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%) 200 2e-51
TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 145 5e-35
TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 145 8e-35
TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete 136 3e-32
TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, parti... 132 7e-31
TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, pa... 130 2e-30
TC220331 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partia... 126 4e-29
TC231559 homologue to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (71%) 123 3e-28
TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%) 122 6e-28
CF922412 120 2e-27
TC227518 similar to UP|Q93VD3 (Q93VD3) At1g30270/F12P21_6 (CBL-i... 120 2e-27
TC207101 similar to UP|O24342 (O24342) Serine/threonine kinase ,... 110 2e-24
TC227273 weakly similar to UP|KGS9_YEAST (P43637) Probable serin... 108 6e-24
TC226920 weakly similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial... 106 4e-23
TC217983 UP|Q8GSL0 (Q8GSL0) Phosphoenolpyruvate carboxylase kina... 105 7e-23
TC230049 similar to UP|Q9MAV2 (Q9MAV2) F24O1.13, partial (47%) 104 1e-22
TC225868 UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, complete 104 2e-22
>TC205120 weakly similar to UP|Q7KSD3 (Q7KSD3) CG7693-PA (Cg7693-pb)
(RE53265p), partial (20%)
Length = 2170
Score = 363 bits (931), Expect = e-100
Identities = 205/462 (44%), Positives = 274/462 (58%), Gaps = 1/462 (0%)
Frame = +2
Query: 129 REVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGT 188
+E LK LEYLH HGHIHRDVKAGN+L+D G VKL DFGVSAC+FD+GDRQRSRNTFVGT
Sbjct: 2 KETLKALEYLHRHGHIHRDVKAGNILLDDNGQVKLADFGVSACMFDTGDRQRSRNTFVGT 181
Query: 189 PCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDY 248
PCW+APEV++ GYNFKADIWSFGITALELAHGHAPFSKYPP+KVLLMT+QNAPPGLDY
Sbjct: 182 PCWIAPEVLQPGTGYNFKADIWSFGITALELAHGHAPFSKYPPMKVLLMTIQNAPPGLDY 361
Query: 249 ERDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDR 308
+RD+KFSKSFK+M+A CLVKD +KRPS KLLKHSFFKQA+ + + L LP L +R
Sbjct: 362 DRDRKFSKSFKEMVAMCLVKDQTKRPSVEKLLKHSFFKQAKPPELSVKKLFADLPPLWNR 541
Query: 309 MEILKRKEDM-LAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSD 367
++ L+ K+ LA KKMP + E +SQ+EY RG+S WNF+++D+KAQA+L+ D DD
Sbjct: 542 VKSLQHKDAAELALKKMPSAEQEAISQSEYHRGVSAWNFDIDDLKAQAALMQDDDD---- 709
Query: 368 ISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSCLDDHVDHS 427
+ + +D+ SS+ + + D + + + TS + + S
Sbjct: 710 ---------IAEMREEDENKFFSSYKGTTDSQFIVDKKNSNNLQQDEFTSQVGSNDIPKS 862
Query: 428 LGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFKH*CLSNFH 487
+V A + + S L + P+ +
Sbjct: 863 EKRNGSVAEATPSTLENDVGTSKVKTQSVKLGKTQSGPLMPGLV--------------LG 1000
Query: 488 HSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTELP 547
HS S F +++ N N L+ N +++ S + +++ + P
Sbjct: 1001HSSSERGRTF---ERFENENQLLGEKSN-----RDIRRAPSFSGPLMLPNRASANSLSAP 1156
Query: 548 SRASKTSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATP 589
++S D DDK+K +VQ +GRF VTSEN+D K P
Sbjct: 1157IKSS--GGFRDSLDDKSKANLVQIKGRFSVTSENLDLVKDIP 1276
>BM527293
Length = 413
Score = 241 bits (616), Expect = 6e-64
Identities = 107/137 (78%), Positives = 125/137 (91%)
Frame = +3
Query: 70 EAQTMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLR 129
E QTM L+DHPNVL++HCSF + HNLWVVMP+M+GGSCLHI+K+ +P+GFEE VIAT+L
Sbjct: 3 EVQTMNLIDHPNVLRAHCSFTAGHNLWVVMPYMAGGSCLHIMKSNYPEGFEEPVIATLLH 182
Query: 130 EVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTP 189
EVLK L YLH HGHIHRDVK+GN+L+DS GAVKL DFGVSAC+FD+GDRQRSRNTFVGTP
Sbjct: 183 EVLKALVYLHAHGHIHRDVKSGNILLDSNGAVKLADFGVSACMFDAGDRQRSRNTFVGTP 362
Query: 190 CWMAPEVMEQLHGYNFK 206
CWMAPEVM+QLHGY+FK
Sbjct: 363 CWMAPEVMQQLHGYDFK 413
>CA953124
Length = 422
Score = 208 bits (530), Expect = 6e-54
Identities = 100/140 (71%), Positives = 117/140 (83%), Gaps = 1/140 (0%)
Frame = +2
Query: 216 ALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQMIACCLVKDPSKRPS 275
ALELAHGHAPFSKYPP+KVLLMTLQNAPPGLDYERDK+FSK+FK+++A CLVKDP KRPS
Sbjct: 2 ALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYERDKRFSKAFKELVATCLVKDPKKRPS 181
Query: 276 ASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILKRKE-DMLAQKKMPDGQMEELSQ 334
+ KLLKH FFKQAR+S Y+ RT+LEGL LGDR +LK K+ D+L Q K ++LSQ
Sbjct: 182 SEKLLKHHFFKQARASKYLARTILEGLAPLGDRFRLLKAKQADLLVQNKALYEDKDQLSQ 361
Query: 335 NEYKRGISGWNFNLEDMKAQ 354
EY RGIS WNFNLED+K+Q
Sbjct: 362 KEYIRGISAWNFNLEDLKSQ 421
>TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%)
Length = 2224
Score = 200 bits (508), Expect = 2e-51
Identities = 129/388 (33%), Positives = 198/388 (50%), Gaps = 21/388 (5%)
Frame = +1
Query: 80 PNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLH 139
P + + + S+++ LW++M +M+GGS ++++ P +E+ IA +LR++L ++YLH
Sbjct: 28 PYITEYYGSYLNQTKLWIIMEYMAGGSVADLIQSGPP--LDEMSIACILRDLLHAVDYLH 201
Query: 140 HHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQ 199
G IHRD+KA N+L+ G VK+ DFGVSA L + R R TFVGTP WMAPEV++
Sbjct: 202 SEGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISR---RKTFVGTPFWMAPEVIQN 372
Query: 200 LHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFK 259
GYN KADIWS GITA+E+A G P + P++VL + + PP L D FS+ K
Sbjct: 373 TDGYNEKADIWSLGITAIEMAKGEPPLADLHPMRVLFIIPRENPPQL----DDHFSRPLK 540
Query: 260 QMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLE---------------GLPA 304
+ ++ CL K P++RPSA +LLK F + AR S ++ + E G
Sbjct: 541 EFVSLCLKKVPAERPSAKELLKDRFIRNARKSSKLSERIRERPKYQIKEDEETPRNGPSG 720
Query: 305 LGDRMEILKRKEDMLAQK-KMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLIN---- 359
+G+ +K D ++ P Q + L +GW+F++ + + N
Sbjct: 721 MGEASGTVKVARDSRGEENNRPSDQGKTLKS-------AGWDFSIGGSQGTGTFRNVSRP 879
Query: 360 -DFDDAMSDISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSS 418
F D ++++SH LT A S +RS + S+ SS
Sbjct: 880 PQFRDKITEVSH----NQLTQRKAPQSGYQGGSVNRSAL------NESLESSFGRDLRVP 1029
Query: 419 CLDDHVDHSLGDMENVGRAAEVVVATHP 446
D+H+D+ L D E G + VV P
Sbjct: 1030HHDEHLDNHLEDDELSGNGSGTVVIRSP 1113
>TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (73%)
Length = 2291
Score = 145 bits (367), Expect = 5e-35
Identities = 85/263 (32%), Positives = 139/263 (52%), Gaps = 6/263 (2%)
Frame = +1
Query: 30 IGQGVSASVHRALCVSFNEIVAIK----ILDFERDNCDLNNISREAQTMVLVDHPNVLKS 85
+G+G V+ ++ AIK + D L +++E + + HPN+++
Sbjct: 634 LGRGTFGHVYLGFNSENGQMCAIKEVKVVFDDHTSKECLKQLNQEINLLNQLSHPNIVQY 813
Query: 86 HCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIH 145
H S + + +L V + ++SGGS +L+ P F+E VI R+++ GL YLH +H
Sbjct: 814 HGSELVEESLSVYLEYVSGGSIHKLLQEYGP--FKEPVIQNYTRQIVSGLAYLHGRNTVH 987
Query: 146 RDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHGYNF 205
RD+K N+L+D G +KL DFG++ + S S +F G+P WMAPEV+ +GY+
Sbjct: 988 RDIKGANILVDPNGEIKLADFGMAKHINSSA----SMLSFKGSPYWMAPEVVMNTNGYSL 1155
Query: 206 KADIWSFGITALELAHGHAPFSKYPPLKVL--LMTLQNAPPGLDYERDKKFSKSFKQMIA 263
DIWS G T +E+A P+++Y + + + ++ P E + S K+ I
Sbjct: 1156PVDIWSLGCTIIEMATSKPPWNQYEGVAAIFKIGNSKDMP-----EIPEHLSNDAKKFIK 1320
Query: 264 CCLVKDPSKRPSASKLLKHSFFK 286
CL +DP RP+A KLL H F +
Sbjct: 1321LCLQRDPLARPTAQKLLDHPFIR 1389
>TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (51%)
Length = 1451
Score = 145 bits (365), Expect = 8e-35
Identities = 84/263 (31%), Positives = 138/263 (51%), Gaps = 6/263 (2%)
Frame = +3
Query: 30 IGQGVSASVHRALCVSFNEIVAIK----ILDFERDNCDLNNISREAQTMVLVDHPNVLKS 85
+G+G V+ ++ AIK + D + L +++E + + HPN+++
Sbjct: 672 LGRGTFGHVYLGFNSDSGQLCAIKEVRVVCDDQSSKECLKQLNQEIHLLSQLSHPNIVQY 851
Query: 86 HCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIH 145
+ S + + L V + ++SGGS +L+ F+E VI R+++ GL YLH +H
Sbjct: 852 YGSDLGEETLSVYLEYVSGGSIHKLLQEY--GAFKEPVIQNYTRQIVSGLSYLHGRNTVH 1025
Query: 146 RDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHGYNF 205
RD+K N+L+D G +KL DFG++ + S S +F G+P WMAPEV+ +GY+
Sbjct: 1026RDIKGANILVDPNGEIKLADFGMAKHINSSS----SMLSFKGSPYWMAPEVVMNTNGYSL 1193
Query: 206 KADIWSFGITALELAHGHAPFSKYPPLKVL--LMTLQNAPPGLDYERDKKFSKSFKQMIA 263
DIWS G T LE+A P+++Y + + ++ P D+ S K I
Sbjct: 1194PVDIWSLGCTILEMATSKPPWNQYEGAAAIFKIGNSRDMPEIPDH-----LSSDAKNFIQ 1358
Query: 264 CCLVKDPSKRPSASKLLKHSFFK 286
CL +DPS RP+A KL++H F +
Sbjct: 1359LCLQRDPSARPTAQKLIEHPFIR 1427
>TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete
Length = 1502
Score = 136 bits (343), Expect = 3e-32
Identities = 87/263 (33%), Positives = 138/263 (52%), Gaps = 8/263 (3%)
Frame = +2
Query: 30 IGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDHPNVLKSHCSF 89
+G+G V ++ A+K++ + I++E + P V+ + SF
Sbjct: 314 VGKGNGGVVQLVQHKWTSQFFALKVIQMNIEESMRKQIAQELKINQQAQCPYVVVCYQSF 493
Query: 90 VSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHI-HRDV 148
+ + +++ +M GGS +LK E +A + ++VLKGL YLHH HI HRD+
Sbjct: 494 YENGVISIILEYMDGGSLADLLKKVKT--IPEDYLAAICKQVLKGLVYLHHEKHIIHRDL 667
Query: 149 KAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPE-VMEQLHGYNFKA 207
K N+LI+ G VK+ DFGVSA + + + NTF+GT +M+PE + GYN+K+
Sbjct: 668 KPSNLLINHIGEVKITDFGVSAIMESTSGQ---ANTFIGTYNYMSPERINGSQRGYNYKS 838
Query: 208 DIWSFGITALELAHGHAPFS------KYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQM 261
DIWS G+ LE A G P++ + + L+ T+ + PP + ++FS F
Sbjct: 839 DIWSLGLILLECALGRFPYAPPDQSETWESIFELIETIVDKPPPI--PPSEQFSTEFCSF 1012
Query: 262 IACCLVKDPSKRPSASKLLKHSF 284
I+ CL KDP R SA +L+ H F
Sbjct: 1013ISACLQKDPKDRLSAQELMAHPF 1081
>TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, partial (37%)
Length = 1866
Score = 132 bits (331), Expect = 7e-31
Identities = 101/377 (26%), Positives = 166/377 (43%), Gaps = 15/377 (3%)
Frame = +1
Query: 30 IGQGVSASVHRALCVSFNEIVAIKILDFERDNCD----LNNISREAQTMVLVDHPNVLKS 85
IG+G SV+ A + A+K +D D+ + + +E + + + HPN+++
Sbjct: 454 IGRGSYGSVYHATNLETGASCALKEVDLFPDDPKSADCIKQLEQEIRILRQLHHPNIVQY 633
Query: 86 HCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIH 145
+ S + L++ M ++ GS LH H E V+ R +L GL YLH IH
Sbjct: 634 YGSEIVGDRLYIYMEYVHPGS-LHKFMHEHCGAMTESVVRNFTRHILSGLAYLHGTKTIH 810
Query: 146 RDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVM------EQ 199
RD+K N+L+D+ G+VKL DFGVS L + + + G+P WMAPE+M E
Sbjct: 811 RDIKGANLLVDASGSVKLADFGVSKILTE----KSYELSLKGSPYWMAPELMKAAIKKES 978
Query: 200 LHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFK 259
DIWS G T +E+ G P+S++ + + L +P + + S +
Sbjct: 979 SPDIAMAIDIWSLGCTIIEMLTGKPPWSEFEGPQAMFKVLHKSP-----DLPESLSSEGQ 1143
Query: 260 QMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYIT----RTLLEGLPALGDRMEILKRK 315
+ C ++P++RPSA+ LL H+F + D + G P GD
Sbjct: 1144DFLQQCFRRNPAERPSAAVLLTHAFVQNLHDQDVQVHSQGQNCPRGDPGPGD-------- 1299
Query: 316 EDMLAQKKMPDGQMEELSQNEYKRGISGWNFN-LEDMKAQASLINDFDDAMSDISHVSSA 374
++ G S+ I W FN ++ S +D+ S S
Sbjct: 1300----DSRRHSPGNSSRNSRGVGSASIRAWFFNKTHNLIGVTSKKYVSEDSNHVTSSRDSP 1467
Query: 375 CSLTNLDAQDKQLPSSS 391
CS+T ++ + SS+
Sbjct: 1468CSMTEMNNPQSPMKSSA 1518
>TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (91%)
Length = 1377
Score = 130 bits (327), Expect = 2e-30
Identities = 89/262 (33%), Positives = 137/262 (51%), Gaps = 7/262 (2%)
Frame = +2
Query: 30 IGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDHPNVLKSHCSF 89
IG G +V++ + + + A+K++ + I RE Q + VD NV+K H +
Sbjct: 299 IGSGSGGTVYKVVHRTSGRVYALKVIYGHHEESVRRQIHREIQILRDVDDANVVKCHEMY 478
Query: 90 VSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIHRDVK 149
+ + V++ FM GGS L+ H +E +A + R++L+GL YLH +HRD+K
Sbjct: 479 DQNSEIQVLLEFMDGGS----LEGKHIT--QEQQLADLSRQILRGLAYLHRRHIVHRDIK 640
Query: 150 AGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPE-VMEQLHGYNFKA- 207
N+LI+SR VK+ DFGV L + D N+ VGT +M+PE + ++ + A
Sbjct: 641 PSNLLINSRKQVKIADFGVGRILNQTMD---PCNSSVGTIAYMSPERINTDINDGQYDAY 811
Query: 208 --DIWSFGITALELAHGHAPFS---KYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQMI 262
DIWSFG++ LE G PF+ + ++ + PP E S FK I
Sbjct: 812 AGDIWSFGVSILEFYMGRFPFAVGRQGDWASLMCAICMSQPP----EAPPSASPHFKDFI 979
Query: 263 ACCLVKDPSKRPSASKLLKHSF 284
CL +DPS+R SAS+LL+H F
Sbjct: 980 LRCLQRDPSRRWSASRLLEHPF 1045
>TC220331 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial (62%)
Length = 669
Score = 126 bits (316), Expect = 4e-29
Identities = 76/215 (35%), Positives = 120/215 (55%), Gaps = 10/215 (4%)
Frame = +1
Query: 80 PNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLH 139
P V+ + SF + + +++ +M GGS +L E ++ + ++VLKGL YLH
Sbjct: 7 PYVVVCYNSFYHNGVISIILEYMDGGSLEDLLSKVKT--IPESYLSAICKQVLKGLMYLH 180
Query: 140 HHGHI-HRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPE-VM 197
+ HI HRD+K N+LI+ RG VK+ DFGVS + ++ + NTF+GT +M+PE ++
Sbjct: 181 YAKHIIHRDLKPSNLLINHRGEVKITDFGVSVIMENTSGQ---ANTFIGTYSYMSPERII 351
Query: 198 EQLHGYNFKADIWSFGITALELAHGHAPFSKYPP--------LKVLLMTLQNAPPGLDYE 249
HGYN+K+DIWS G+ L+ A G P++ PP +++ + ++ P +
Sbjct: 352 GNQHGYNYKSDIWSLGLILLKCATGQFPYT--PPDREGWENIFQLIEVIVEKPSPSAPSD 525
Query: 250 RDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSF 284
FS F I+ CL K+P RPSA L+ H F
Sbjct: 526 ---DFSPEFCSFISACLQKNPGDRPSARDLINHPF 621
>TC231559 homologue to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (71%)
Length = 867
Score = 123 bits (308), Expect = 3e-28
Identities = 78/220 (35%), Positives = 118/220 (53%), Gaps = 7/220 (3%)
Frame = +2
Query: 80 PNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLH 139
P+V+ + SF + + +V+ +M GS ++K E +A V ++VL+GL YLH
Sbjct: 80 PHVVVCYHSFYHNGVISLVLEYMDRGSLADVIKQVKT--ILEPYLAVVFKQVLQGLVYLH 253
Query: 140 HHGH-IHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVME 198
+ H IHRD+K N+L++ +G VK+ DFGVSA L S R+TFVGT +M+PE +
Sbjct: 254 NERHVIHRDIKPSNLLVNHKGEVKITDFGVSAMLASS---MGQRDTFVGTYNYMSPERIS 424
Query: 199 QLHGYNFKADIWSFGITALELAHGHAPF------SKYPPLKVLLMTLQNAPPGLDYERDK 252
Y++ +DIWS G+ LE A G P+ +P LL + +PP
Sbjct: 425 G-STYDYSSDIWSLGMVVLECAIGRFPYIQSEDQQSWPSFYELLAAIVESPP--PSAPPD 595
Query: 253 KFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSD 292
+FS F ++ C+ KDP R ++ KLL H F K+ D
Sbjct: 596 QFSPEFCSFVSSCIQKDPRDRLTSLKLLDHPFIKKFEDKD 715
>TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%)
Length = 1190
Score = 122 bits (306), Expect = 6e-28
Identities = 87/265 (32%), Positives = 137/265 (50%), Gaps = 10/265 (3%)
Frame = +1
Query: 30 IGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLV-DHPNVLKSHCS 88
+G G +V++ + + A+KI+ + D E + V D P+V++ H S
Sbjct: 235 LGHGNGGTVYKVRHKATSATYALKIIHSDTDATRRRRALSETSILRRVTDCPHVVRFHSS 414
Query: 89 FVSDH-NLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIHRD 147
F ++ ++M +M GG+ L A+ F E +A V R+VL+GL YLH HRD
Sbjct: 415 FEKPSGDVAILMEYMDGGTLETALAASGT--FSEERLAKVARDVLEGLAYLHARNIAHRD 588
Query: 148 VKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVME-QLHGYN-- 204
+K N+L++S G VK+ DFGVS + S + N++VGT +M+P+ + + +G N
Sbjct: 589 IKPANILVNSEGDVKIADFGVSKLMCRS---LEACNSYVGTCAYMSPDRFDPEAYGGNYN 759
Query: 205 -FKADIWSFGITALELAHGHAPF---SKYPPLKVLLMTL-QNAPPGLDYERDKKFSKSFK 259
F ADIWS G+T EL GH PF + P L+ + PP L + S F+
Sbjct: 760 GFAADIWSLGLTLFELYVGHFPFLQAGQRPDWATLMCAICFGDPPSL----PETASPEFR 927
Query: 260 QMIACCLVKDPSKRPSASKLLKHSF 284
+ CCL K+ +R + ++LL H F
Sbjct: 928 DFVECCLKKESGERWTTAQLLTHPF 1002
>CF922412
Length = 701
Score = 120 bits (302), Expect = 2e-27
Identities = 72/191 (37%), Positives = 109/191 (56%), Gaps = 1/191 (0%)
Frame = -2
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFER-DNCDLNNISREAQTMVLVDHPNV 82
Y L +EIG+G V++ L + + VAIK + E DLN I +E + ++H N+
Sbjct: 562 YMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMQEIDLLKNLNHKNI 383
Query: 83 LKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHG 142
+K S + +L +V+ ++ GS +I+K F E ++A + +VL+GL YLH G
Sbjct: 382 VKYLGSSKTKSHLHIVLEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQG 203
Query: 143 HIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHG 202
IHRD+K N+L G VKL DFGV+ L ++ + ++ VGTP WMAPEV+E + G
Sbjct: 202 VIHRDIKGANILTTKEGLVKLADFGVATKLTEA---DVNTHSVVGTPYWMAPEVIE-MAG 35
Query: 203 YNFKADIWSFG 213
+DIWS G
Sbjct: 34 VCAASDIWSVG 2
>TC227518 similar to UP|Q93VD3 (Q93VD3) At1g30270/F12P21_6 (CBL-interacting
protein kinase 23), partial (86%)
Length = 1492
Score = 120 bits (301), Expect = 2e-27
Identities = 84/266 (31%), Positives = 138/266 (51%), Gaps = 2/266 (0%)
Frame = +1
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFER--DNCDLNNISREAQTMVLVDHPN 81
Y+L +G+G A V A V E VAIKILD E+ + + I RE TM L+ HPN
Sbjct: 37 YELGRTLGEGNFAKVKFARHVETRENVAIKILDKEKLLKHKMIAQIKREISTMKLIRHPN 216
Query: 82 VLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHH 141
V++ + S +++V+ F++GG K A +E ++++ ++Y H
Sbjct: 217 VIRMYEVMASKTKIYIVLEFVTGGELFD--KIARSGRLKEDEARKYFQQLICAVDYCHSR 390
Query: 142 GHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLH 201
G HRD+K N+L+D+ G +K+ DFG+SA L +T GTP ++APEV+
Sbjct: 391 GVFHRDLKPENLLLDANGVLKVSDFGLSA-LPQQVREDGLLHTTCGTPNYVAPEVINNKG 567
Query: 202 GYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQM 261
KAD+WS G+ L G+ PF + L L + A ++ FS S K++
Sbjct: 568 YDGAKADLWSCGVILFVLMAGYLPFEE-TNLSALYKKIFKA----EFTCPPWFSSSAKKL 732
Query: 262 IACCLVKDPSKRPSASKLLKHSFFKQ 287
I L +P+ R + ++++++ +FK+
Sbjct: 733 INKILDPNPATRITFAEVIENDWFKK 810
>TC207101 similar to UP|O24342 (O24342) Serine/threonine kinase , partial
(98%)
Length = 1719
Score = 110 bits (275), Expect = 2e-24
Identities = 82/270 (30%), Positives = 131/270 (48%), Gaps = 6/270 (2%)
Frame = +1
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFER--DNCDLNNISREAQTMVLVDHPN 81
Y++ IG+G A V A E VA+KILD E+ + I RE TM L+ HPN
Sbjct: 220 YEVGRTIGEGTFAKVKFARNSETGEPVALKILDKEKVLKHKMAEQIRREVATMKLIKHPN 399
Query: 82 VLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHH 141
V++ + S +++V+ F++GG K + E ++++ ++Y H
Sbjct: 400 VVRLYEVMGSKTKIYIVLEFVTGGELFD--KIVNHGRMSENEARRYFQQLINAVDYCHSR 573
Query: 142 GHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLH 201
G HRD+K N+L+D+ G +K+ DFG+SA D +T GTP ++APEV+
Sbjct: 574 GVYHRDLKPENLLLDTYGNLKVSDFGLSALSQQVRD-DGLLHTTCGTPNYVAPEVLNDRG 750
Query: 202 GYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNA----PPGLDYERDKKFSKS 257
AD+WS G+ L G+ PF P L L + A PP L + +
Sbjct: 751 YDGATADLWSCGVILFVLVAGYLPFDD-PNLMNLYKKISAAEFTCPPWLSF--------T 903
Query: 258 FKQMIACCLVKDPSKRPSASKLLKHSFFKQ 287
+++I L DP+ R + ++L +FK+
Sbjct: 904 ARKLITRILDPDPTTRITIPEILDDEWFKK 993
>TC227273 weakly similar to UP|KGS9_YEAST (P43637) Probable
serine/threonine-protein kinase YGL179C , partial (8%)
Length = 2373
Score = 108 bits (271), Expect = 6e-24
Identities = 88/291 (30%), Positives = 135/291 (46%), Gaps = 26/291 (8%)
Frame = +3
Query: 17 YPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCD-LNNISREAQTMV 75
+ I + ++G G ++R S + VAIK+L ER + D L ++E M
Sbjct: 990 WEIDTNQLKYENKVGSGSFGDLYRGTYCS--QDVAIKVLKPERISTDMLREFAQEVYIMR 1163
Query: 76 LVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGL 135
+ H NV++ + NL +V FMS GS L F+ + V +V KG+
Sbjct: 1164 KIRHKNVVQFIGACTRPPNLCIVTEFMSRGSLYDFLHKQR-GVFKLPSLLKVAIDVSKGM 1340
Query: 136 EYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPE 195
YLH + IHRD+K N+L+D VK+ DFGV+ SG GT WMAPE
Sbjct: 1341 NYLHQNNIIHRDLKTANLLMDENEVVKVADFGVARVQTQSG----VMTAETGTYRWMAPE 1508
Query: 196 VMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQ---------NAPPGL 246
V+E Y+ KAD++SFGI EL G P+S PL+ + +Q N P L
Sbjct: 1509 VIEH-KPYDQKADVFSFGIALWELLTGELPYSCLTPLQAAVGVVQKGLRPTIPKNTHPRL 1685
Query: 247 D---YERD-------------KKFSKSFKQMIACCLVKDPSKRPSASKLLK 281
Y++ K +++ C +DP++RP+ S++++
Sbjct: 1686 S*LHYKQQLAWCRRDQLSTIPKNTHPRLSELLQRCWQQDPTQRPNFSEVIE 1838
>TC226920 weakly similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (49%)
Length = 722
Score = 106 bits (264), Expect = 4e-23
Identities = 70/185 (37%), Positives = 101/185 (53%), Gaps = 9/185 (4%)
Frame = +2
Query: 109 HILKAAHPDG-FEEVVIATVLREVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFG 167
H L A G F E +A V R+VL+GL YLH HRD+K N+L++S G VK+ DFG
Sbjct: 11 HPLTALATGGTFSEERLAKVARDVLEGLAYLHARNIAHRDIKPANILVNSEGEVKIADFG 190
Query: 168 VSACLFDSGDRQRSRNTFVGTPCWMAPEVME-QLHGYN---FKADIWSFGITALELAHGH 223
VS + + + N++VGT +M+P+ + + +G N F ADIWS G+T EL GH
Sbjct: 191 VSKLMCRT---LEACNSYVGTCAYMSPDRFDPEAYGGNYNGFAADIWSLGLTLFELYVGH 361
Query: 224 APF---SKYPPLKVLLMTL-QNAPPGLDYERDKKFSKSFKQMIACCLVKDPSKRPSASKL 279
PF + P L+ + + PP L + S F + CCL K+ +R +A++L
Sbjct: 362 FPFLQAGQRPDWATLMCAICFSDPPSL----PETASPEFHDFVECCLKKESGERWTAAQL 529
Query: 280 LKHSF 284
L H F
Sbjct: 530 LTHPF 544
>TC217983 UP|Q8GSL0 (Q8GSL0) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1203
Score = 105 bits (262), Expect = 7e-23
Identities = 89/298 (29%), Positives = 143/298 (47%), Gaps = 4/298 (1%)
Frame = +3
Query: 2 KDKDRDKEKETEKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFE--R 59
K+K R++++ K +YQL EEIG+G ++ R NE A K++D
Sbjct: 9 KEKRRNEDRMCTALK-----TNYQLSEEIGRGRFGTIFRCFHPLSNEPYACKLIDKSLLH 173
Query: 60 DNCDLNNISREAQTMVLVD-HPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDG 118
D+ D + + E + M L+ HPN+L+ F D L +VM H L DG
Sbjct: 174 DSTDRDCLQNEPKFMTLLSPHPNILQIFHVFEDDQYLSIVMDLCQP----HTLFDRMVDG 341
Query: 119 -FEEVVIATVLREVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGD 177
+E A +++ +L+ + + H G HRD+K N+L DS +KL DFG SA F G
Sbjct: 342 PIQESQAAALMKNLLEAVAHCHRLGVAHRDIKPDNILFDSADNLKLADFG-SAEWFGDG- 515
Query: 178 RQRSRNTFVGTPCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLM 237
RS + VGTP ++APEV+ Y+ K D+WS G+ + G PF ++
Sbjct: 516 --RSMSGVVGTPYYVAPEVLLG-REYDEKVDVWSCGVILYIMLAGIPPFYGDSAAEIFEA 686
Query: 238 TLQNAPPGLDYERDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYIT 295
++ A + S + K ++ + +D S+R SA + L+H + A + +T
Sbjct: 687 VVR-ANLRFPSRIFRTVSPAAKDLLRKMICRDSSRRFSAEQALRHPWILSAGDTAELT 857
>TC230049 similar to UP|Q9MAV2 (Q9MAV2) F24O1.13, partial (47%)
Length = 1216
Score = 104 bits (260), Expect = 1e-22
Identities = 75/277 (27%), Positives = 127/277 (45%), Gaps = 18/277 (6%)
Frame = +2
Query: 50 VAIKILDFERDNCDL-----NNISREAQTMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSG 104
VAIK++ ++ DL + E ++ + HPN++ + ++ +++G
Sbjct: 236 VAIKLISQPEEDEDLAAFLEKQFTSEVSLLLRLGHPNIITFIAACKKPPVFCIITEYLAG 415
Query: 105 GSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLG 164
GS L P+ ++ + ++ +G++YLH G +HRD+K+ N+L+ VK+
Sbjct: 416 GSLGKFLHHQQPNILPLKLVLKLALDIARGMKYLHSQGILHRDLKSENLLLGEDMCVKVA 595
Query: 165 DFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHA 224
DFG+S G S F GT WMAPE++++ H + K D++SFGI EL G
Sbjct: 596 DFGISCLESQCG----SAKGFTGTYRWMAPEMIKEKH-HTKKVDVYSFGIVLWELLTGKT 760
Query: 225 PFSKYPPLK-VLLMTLQNAPPGLDYERDKKFSKSFKQMIACCLVKDPSKRPSASKLLK-- 281
PF P + ++ +NA P L K +F +I C +P KRP +++
Sbjct: 761 PFDNMTPEQAAYAVSHKNARPPL----PSKCPWAFSDLINRCWSSNPDKRPHFDEIVSIL 928
Query: 282 ----------HSFFKQARSSDYITRTLLEGLPALGDR 308
FF + S + T+L LP R
Sbjct: 929 EYYTESLQQDPEFFSTYKPSPTSSNTILGCLPKCNAR 1039
>TC225868 UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, complete
Length = 2261
Score = 104 bits (259), Expect = 2e-22
Identities = 75/267 (28%), Positives = 128/267 (47%), Gaps = 3/267 (1%)
Frame = +2
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFER--DNCDLNNISREAQTMVLVDHPN 81
Y+L +G G A V+ A + + VA+K++ E+ + I RE M +V HPN
Sbjct: 488 YELGRLLGHGTFAKVYHARHLKTGKSVAMKVVGKEKVVKVGMMEQIKREISAMNMVKHPN 667
Query: 82 VLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHH 141
+++ H S +++ M + GG + + E + ++++ +++ H
Sbjct: 668 IVQLHEVMASKSKIYIAMELVRGGELFNKIARGR---LREEMARLYFQQLISAVDFCHSR 838
Query: 142 GHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRS-RNTFVGTPCWMAPEVMEQL 200
G HRD+K N+L+D G +K+ DFG+S F R +T GTP ++APEV+ +
Sbjct: 839 GVYHRDLKPENLLLDDDGNLKVTDFGLST--FSEHLRHDGLLHTTCGTPAYVAPEVIGKR 1012
Query: 201 HGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQ 260
KADIWS G+ L G PF L+ L D++ FS ++
Sbjct: 1013GYDGAKADIWSCGVILYVLLAGFLPFQDDN-----LVALYKKIYRGDFKCPPWFSSEARR 1177
Query: 261 MIACCLVKDPSKRPSASKLLKHSFFKQ 287
+I L +P+ R + SK++ S+FK+
Sbjct: 1178LITKLLDPNPNTRITISKIMDSSWFKK 1258
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,115,418
Number of Sequences: 63676
Number of extensions: 516185
Number of successful extensions: 4345
Number of sequences better than 10.0: 692
Number of HSP's better than 10.0 without gapping: 3761
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3913
length of query: 718
length of database: 12,639,632
effective HSP length: 104
effective length of query: 614
effective length of database: 6,017,328
effective search space: 3694639392
effective search space used: 3694639392
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144503.1


