

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144484.9 + phase: 0
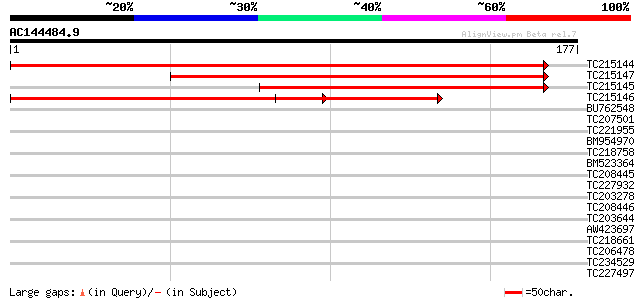
(177 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC215144 similar to UP|ATPQ_ARATH (Q9FT52) ATP synthase D chain,... 313 2e-86
TC215147 similar to GB|CAC81059.1|17939851|ATH271469 mitochondri... 174 1e-44
TC215145 similar to GB|CAC81059.1|17939851|ATH271469 mitochondri... 169 6e-43
TC215146 similar to UP|ATPQ_ARATH (Q9FT52) ATP synthase D chain,... 163 3e-41
BU762548 similar to PIR|D86292|D862 protein F7H2.14 [imported] -... 32 0.20
TC207501 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F2... 31 0.35
TC221955 weakly similar to UP|Q6UAL1 (Q6UAL1) Myosin heavy chain... 30 0.78
BM954970 similar to GP|17979500|gb| At1g80610/T21F11_6 {Arabidop... 30 0.78
TC218758 similar to UP|Q9FFB5 (Q9FFB5) Emb|CAB89309.1, partial (... 29 1.0
BM523364 homologue to PIR|A96583|A965 F15I1.28 [imported] - Arab... 28 1.7
TC208445 similar to UP|Q876D1 (Q876D1) MNN4 (Fragment), partial ... 28 1.7
TC227932 28 1.7
TC203278 similar to UP|Q9M5A8 (Q9M5A8) Chaperonin 21 precursor, ... 28 2.3
TC208446 similar to UP|MNN4_YEAST (P36044) MNN4 protein, partial... 28 2.3
TC203644 similar to UP|Q9M5A8 (Q9M5A8) Chaperonin 21 precursor, ... 28 2.3
AW423697 28 3.0
TC218661 similar to UP|Q8W437 (Q8W437) PBng143, partial (74%) 27 3.9
TC206478 weakly similar to UP|Q8VYI7 (Q8VYI7) At1g80610/T21F11_6... 27 3.9
TC234529 27 5.0
TC227497 weakly similar to UP|MID2_YEAST (P36027) Mating process... 27 5.0
>TC215144 similar to UP|ATPQ_ARATH (Q9FT52) ATP synthase D chain,
mitochondrial , complete
Length = 843
Score = 313 bits (803), Expect = 2e-86
Identities = 149/168 (88%), Positives = 161/168 (95%)
Frame = +3
Query: 1 MSGTVKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPE 60
MSGTVKKV+DVAFK G+ IDWDGMAKLLV+DEARREF NLRRAFDEVN+QL+TKFSQEPE
Sbjct: 69 MSGTVKKVSDVAFKVGRNIDWDGMAKLLVSDEARREFSNLRRAFDEVNSQLQTKFSQEPE 248
Query: 61 PIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKE 120
PIDW+YYRKGIGTRLVDMYK+HYESIEIPKFVDTVTPQYKPKFDALL+ELKEAEEKSLKE
Sbjct: 249 PIDWDYYRKGIGTRLVDMYKEHYESIEIPKFVDTVTPQYKPKFDALLIELKEAEEKSLKE 428
Query: 121 SERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
SERLEKEI VQ LKK++STMTADEYF +HPEL+KKFDDEIRNDNWGY
Sbjct: 429 SERLEKEIAEVQELKKKLSTMTADEYFEKHPELRKKFDDEIRNDNWGY 572
>TC215147 similar to GB|CAC81059.1|17939851|ATH271469 mitochondrial F0 ATP
synthase D chain {Arabidopsis thaliana;} , partial (81%)
Length = 443
Score = 174 bits (442), Expect = 1e-44
Identities = 83/118 (70%), Positives = 98/118 (82%)
Frame = +1
Query: 51 LETKFSQEPEPIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVEL 110
L T+F+QEP P+DW++ +KGIG L D K+ YES+EIP+F TP Y P FDALL+E+
Sbjct: 10 LPTQFNQEP*PLDWDFXKKGIGPCLXDXXKEPYESLEIPQFGXXGTPPY*P*FDALLIEV 189
Query: 111 KEAEEKSLKESERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
KEAEEKSLKESERLEKEI VQ LKK++STMTADEYF +HPEL+KKFDDEIRNDNWGY
Sbjct: 190 KEAEEKSLKESERLEKEIAEVQELKKKLSTMTADEYFEKHPELRKKFDDEIRNDNWGY 363
>TC215145 similar to GB|CAC81059.1|17939851|ATH271469 mitochondrial F0 ATP
synthase D chain {Arabidopsis thaliana;} , partial (62%)
Length = 795
Score = 169 bits (428), Expect = 6e-43
Identities = 79/90 (87%), Positives = 86/90 (94%)
Frame = +1
Query: 79 YKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRI 138
YK+HYESIE+PKFVDTVTPQYK KFDALL+ELKEAEEKSLKESERLEKEIV VQ LKK++
Sbjct: 10 YKEHYESIEVPKFVDTVTPQYKTKFDALLIELKEAEEKSLKESERLEKEIVEVQELKKKL 189
Query: 139 STMTADEYFAEHPELKKKFDDEIRNDNWGY 168
STMTADEYF +HPEL+KKFDDEIRNDNWGY
Sbjct: 190 STMTADEYFEKHPELRKKFDDEIRNDNWGY 279
>TC215146 similar to UP|ATPQ_ARATH (Q9FT52) ATP synthase D chain,
mitochondrial , partial (79%)
Length = 670
Score = 163 bits (413), Expect = 3e-41
Identities = 77/99 (77%), Positives = 88/99 (88%)
Frame = +2
Query: 1 MSGTVKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPE 60
MSGTVKKV+DVAFK G+ IDWDGMAKLLV+DEARREF NLRRAFDEVN+QL+TKFSQEPE
Sbjct: 44 MSGTVKKVSDVAFKVGRNIDWDGMAKLLVSDEARREFSNLRRAFDEVNSQLQTKFSQEPE 223
Query: 61 PIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQY 99
PIDW+YYRKGIGTRLVDMYK+HYES+ + V+ + Y
Sbjct: 224 PIDWDYYRKGIGTRLVDMYKEHYESMFLNSVVNALNIVY 340
Score = 93.2 bits (230), Expect = 6e-20
Identities = 46/52 (88%), Positives = 48/52 (91%)
Frame = +3
Query: 84 ESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLK 135
+ IEIPKFVDTVTPQYKPKFDALL+ELKEAEEKSLKESERLEKEI VQ LK
Sbjct: 417 KGIEIPKFVDTVTPQYKPKFDALLIELKEAEEKSLKESERLEKEIAEVQELK 572
>BU762548 similar to PIR|D86292|D862 protein F7H2.14 [imported] - Arabidopsis
thaliana, partial (8%)
Length = 438
Score = 31.6 bits (70), Expect = 0.20
Identities = 23/84 (27%), Positives = 44/84 (52%), Gaps = 1/84 (1%)
Frame = +3
Query: 83 YESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRISTMT 142
++ +++ +T T + K + L ELKE E+ LKE LEKE+ +++ +T
Sbjct: 78 FDILQVANPSETATTR-KTRRKKTLAELKEEEDLLLKERRNLEKELASLR--------LT 230
Query: 143 ADEYFAEHPELKK-KFDDEIRNDN 165
++ A + +K+ K D E R ++
Sbjct: 231 IQKHRATNESIKRLKLDFESRQNS 302
>TC207501 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F28A23_50
{Arabidopsis thaliana;} , partial (52%)
Length = 724
Score = 30.8 bits (68), Expect = 0.35
Identities = 16/40 (40%), Positives = 28/40 (70%)
Frame = -2
Query: 102 KFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRISTM 141
+++++LVE K+ ++K LKE RL++ V LK ++STM
Sbjct: 174 RWESILVE*KDEQQKRLKEKHRLQQ----VP*LKSQVSTM 67
>TC221955 weakly similar to UP|Q6UAL1 (Q6UAL1) Myosin heavy chain class XI E3
protein, partial (6%)
Length = 728
Score = 29.6 bits (65), Expect = 0.78
Identities = 22/79 (27%), Positives = 39/79 (48%), Gaps = 6/79 (7%)
Frame = +1
Query: 105 ALLVELKEAEEKSLKESERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDD----- 159
A ++ +EA + +++++ + KE+ V K + T +E E ELKKK +
Sbjct: 67 AAIIHEREAAKIAIEQAPPVIKEVPVVDETKLELLTNKNEELETEVEELKKKIKEFEESY 246
Query: 160 -EIRNDNWGYSHYKEEQNA 177
EI N+N + KE + A
Sbjct: 247 SEIENEN--QARLKEAEEA 297
>BM954970 similar to GP|17979500|gb| At1g80610/T21F11_6 {Arabidopsis
thaliana}, partial (22%)
Length = 422
Score = 29.6 bits (65), Expect = 0.78
Identities = 25/96 (26%), Positives = 43/96 (44%), Gaps = 4/96 (4%)
Frame = +3
Query: 41 RRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTRLVDMYKQHYES----IEIPKFVDTVT 96
+R + + E+ + P+ W G L D Y++ S ++ +T T
Sbjct: 138 QRRSRSAHNKAESTRASPTTPLSWSGATSASGGNL-DGYEESKPSQTSRSKVANPSETAT 314
Query: 97 PQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQ 132
+ K + L ELKE E+ LKE LEKE+ +++
Sbjct: 315 TR-KTRRKKTLTELKEEEDLLLKERRNLEKELASLR 419
>TC218758 similar to UP|Q9FFB5 (Q9FFB5) Emb|CAB89309.1, partial (59%)
Length = 831
Score = 29.3 bits (64), Expect = 1.0
Identities = 21/83 (25%), Positives = 43/83 (51%), Gaps = 2/83 (2%)
Frame = +1
Query: 47 VNTQLETKFSQEPEPIDWEYYRKGIGTRLVDMYKQH--YESIEIPKFVDTVTPQYKPKFD 104
V T+LE +Q + W YY +G + ++ +Q Y ++E+ + V + K K D
Sbjct: 112 VETKLEKMDNQRSPLLSWAYYFQG---KSMEELRQSLIYTTLELEQTRVAVQEELK-KRD 279
Query: 105 ALLVELKEAEEKSLKESERLEKE 127
L+ LK+ K+++E + +++
Sbjct: 280 EQLLNLKDLLSKTIRERDEAQEK 348
>BM523364 homologue to PIR|A96583|A965 F15I1.28 [imported] - Arabidopsis
thaliana, partial (22%)
Length = 430
Score = 28.5 bits (62), Expect = 1.7
Identities = 11/26 (42%), Positives = 21/26 (80%)
Frame = -2
Query: 112 EAEEKSLKESERLEKEIVNVQSLKKR 137
+++ KS +ESER++ +I+N+ SLK +
Sbjct: 93 KSQTKSDRESERMKTKIINISSLKSQ 16
>TC208445 similar to UP|Q876D1 (Q876D1) MNN4 (Fragment), partial (9%)
Length = 1040
Score = 28.5 bits (62), Expect = 1.7
Identities = 21/62 (33%), Positives = 34/62 (53%), Gaps = 1/62 (1%)
Frame = +1
Query: 80 KQHYESIEIPKFVDTVTPQY-KPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRI 138
K+ + S+ I VD V + K K D + +EAE+K +E ERL+K + +V K +
Sbjct: 232 KKKFSSVTILTVVDLVKEEEAKKKKDEEEKKKEEAEKKRKEEEERLKKMLRSVLCKKCKS 411
Query: 139 ST 140
S+
Sbjct: 412 SS 417
>TC227932
Length = 1562
Score = 28.5 bits (62), Expect = 1.7
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = +1
Query: 35 REFFNLRRAFDEVNTQLETKFSQEPEP 61
+E F+ + FD VN L KFS PEP
Sbjct: 955 KEKFHRKLIFDSVNEILGAKFSSSPEP 1035
>TC203278 similar to UP|Q9M5A8 (Q9M5A8) Chaperonin 21 precursor, partial
(86%)
Length = 1099
Score = 28.1 bits (61), Expect = 2.3
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 3/33 (9%)
Frame = +1
Query: 88 IPKFVDTVTPQY---KPKFDALLVELKEAEEKS 117
+ K V P+Y KP D +L+++KEAEEK+
Sbjct: 265 VVKAATVVAPKYTAIKPLGDRVLIKIKEAEEKT 363
>TC208446 similar to UP|MNN4_YEAST (P36044) MNN4 protein, partial (3%)
Length = 565
Score = 28.1 bits (61), Expect = 2.3
Identities = 21/68 (30%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Frame = +2
Query: 75 LVDMYKQHYESIEIPKFVDTVTPQY--KPKFDALLVELKEAEEKSLKESERLEKEIVNVQ 132
L + K+ + S+ I VD V + K K + + +EAE+K +E ERL+K + +V
Sbjct: 194 LANQLKKKFSSVTILTVVDLVKEEEAKKKKDEEEKKKKEEAEKKRKEEEERLKKILRSVL 373
Query: 133 SLKKRIST 140
K + S+
Sbjct: 374 CKKCKSSS 397
>TC203644 similar to UP|Q9M5A8 (Q9M5A8) Chaperonin 21 precursor, partial
(87%)
Length = 1177
Score = 28.1 bits (61), Expect = 2.3
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 3/33 (9%)
Frame = +3
Query: 88 IPKFVDTVTPQY---KPKFDALLVELKEAEEKS 117
+ K V P+Y KP D +L+++KEAEEK+
Sbjct: 294 VVKAATVVAPKYTAIKPLGDRVLIKIKEAEEKT 392
>AW423697
Length = 417
Score = 27.7 bits (60), Expect = 3.0
Identities = 21/79 (26%), Positives = 37/79 (46%)
Frame = +1
Query: 86 IEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRISTMTADE 145
++ K VD V + K + + E K+ EEK +E +R EKE + + + + E
Sbjct: 160 VDPAKLVDHVYKRTKKQASIVKDEEKKEEEKK-EEEKREEKEEEKKEGEEDNKTEIKRSE 336
Query: 146 YFAEHPELKKKFDDEIRND 164
Y+ + +D EI +D
Sbjct: 337 YWPSKNYIDYAYDPEIFSD 393
>TC218661 similar to UP|Q8W437 (Q8W437) PBng143, partial (74%)
Length = 913
Score = 27.3 bits (59), Expect = 3.9
Identities = 21/67 (31%), Positives = 31/67 (45%), Gaps = 6/67 (8%)
Frame = +3
Query: 42 RAFDEVNTQLE-TKFSQEPEPIDWEYYRKGIGT-----RLVDMYKQHYESIEIPKFVDTV 95
RA EV + E +K Q P DW+ G R V+M ++ +S KFVD
Sbjct: 201 RAISEVAVEPEISKKEQNESPKDWKIKMLYDGDCPLCMREVNMLRERNKSYGTIKFVDIG 380
Query: 96 TPQYKPK 102
+ +Y P+
Sbjct: 381 SDEYSPE 401
>TC206478 weakly similar to UP|Q8VYI7 (Q8VYI7) At1g80610/T21F11_6, partial
(30%)
Length = 1361
Score = 27.3 bits (59), Expect = 3.9
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = +3
Query: 109 ELKEAEEKSLKESERLEKEIVNVQS 133
ELKE E LKES L+KEI +V +
Sbjct: 570 ELKEEESSLLKESIYLKKEIASVNA 644
>TC234529
Length = 504
Score = 26.9 bits (58), Expect = 5.0
Identities = 18/71 (25%), Positives = 30/71 (41%)
Frame = +3
Query: 27 LLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTRLVDMYKQHYESI 86
+LVT E + + +L A ++ + E + W Y+ G L+ +YK HY
Sbjct: 258 VLVTGEE*KNYAHLNFALGKLPLKREKAYITPDMKKQWVYFYLGNKCCLLALYKFHYFFS 437
Query: 87 EIPKFVDTVTP 97
+ P V P
Sbjct: 438 DCPMEVKQGVP 470
>TC227497 weakly similar to UP|MID2_YEAST (P36027) Mating process protein
MID2 (Serine-rich protein SMS1) (Protein kinase A
interference protein), partial (10%)
Length = 742
Score = 26.9 bits (58), Expect = 5.0
Identities = 22/78 (28%), Positives = 30/78 (38%), Gaps = 4/78 (5%)
Frame = +1
Query: 82 HYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNV----QSLKKR 137
H I IP + +TP PKF+ ++ L E + E NV SLKK+
Sbjct: 46 HRFFIHIPLTQNQITPPQNPKFNFSILPLAAVVEMGSRRDEAPAINSTNVFAALGSLKKK 225
Query: 138 ISTMTADEYFAEHPELKK 155
D P+ KK
Sbjct: 226 KKKPEKDHQGPSKPKDKK 279
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,929,846
Number of Sequences: 63676
Number of extensions: 59109
Number of successful extensions: 283
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 282
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 283
length of query: 177
length of database: 12,639,632
effective HSP length: 91
effective length of query: 86
effective length of database: 6,845,116
effective search space: 588679976
effective search space used: 588679976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC144484.9


