

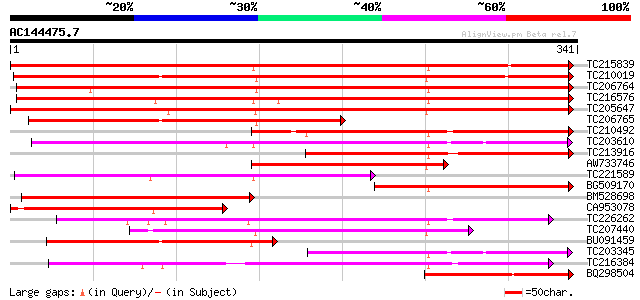
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144475.7 - phase: 0
(341 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC215839 weakly similar to UP|Q84KK5 (Q84KK5) S-adenosyl-L-methi... 367 e-102
TC210019 similar to UP|Q84KK5 (Q84KK5) S-adenosyl-L-methionine: ... 363 e-101
TC206764 similar to UP|7MT9_MEDSA (O22309) Isoflavone-7-O-methyt... 347 4e-96
TC216576 similar to UP|Q84KK6 (Q84KK6) S-adenosyl-L-methionine: ... 311 3e-85
TC205647 similar to UP|Q8GU23 (Q8GU23) Orcinol O-methyltransfera... 285 2e-77
TC206765 similar to UP|Q84KK5 (Q84KK5) S-adenosyl-L-methionine: ... 187 8e-48
TC210492 weakly similar to UP|Q8GU23 (Q8GU23) Orcinol O-methyltr... 172 3e-43
TC203610 weakly similar to UP|Q7X9I9 (Q7X9I9) Caffeic acid O-met... 159 2e-39
TC213916 similar to UP|Q8GU23 (Q8GU23) Orcinol O-methyltransfera... 159 2e-39
AW733746 similar to GP|28804594|db S-adenosyl-L-methionine: daid... 154 7e-38
TC221589 weakly similar to UP|O65859 (O65859) O-methyltransferas... 142 2e-34
BG509170 similar to GP|28804594|db S-adenosyl-L-methionine: daid... 142 2e-34
BM528698 similar to GP|28804594|db S-adenosyl-L-methionine: daid... 137 7e-33
CA953078 weakly similar to GP|28804592|dbj S-adenosyl-L-methioni... 129 1e-30
TC226262 similar to PRF|2119166A.0|1582580|2119166A caffeic acid... 123 1e-28
TC207440 weakly similar to UP|Q8GU21 (Q8GU21) Orcinol O-methyltr... 122 2e-28
BU091459 weakly similar to GP|28804594|db S-adenosyl-L-methionin... 119 3e-27
TC203345 weakly similar to UP|Q7X9I9 (Q7X9I9) Caffeic acid O-met... 116 2e-26
TC216384 weakly similar to PRF|1906376A.0|444327|1906376A O-meth... 114 8e-26
BQ298504 107 6e-24
>TC215839 weakly similar to UP|Q84KK5 (Q84KK5) S-adenosyl-L-methionine:
daidzein 7-0-methyltransferase, partial (55%)
Length = 1369
Score = 367 bits (942), Expect = e-102
Identities = 191/353 (54%), Positives = 245/353 (69%), Gaps = 14/353 (3%)
Frame = +3
Query: 1 MDSENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSAL 60
M S N K ELF Q+ LY Q++ +RP+ +KWAV+LGIPDIIQNH KPI+L +LVS L
Sbjct: 27 MASMNNQKEIELFEGQSLLYMQLYGHLRPMCLKWAVQLGIPDIIQNHAKPISLSDLVSTL 206
Query: 61 RIPEAKAGCVHSVMRLLAHNKIFAIVKIDDNKE-AYALSPTSKLLVKGTDHCLTSMVKMV 119
+IP A A V MR LAHN IF I + ++ E YAL+P SKLLV +DHCL+ MV
Sbjct: 207 QIPPANAAFVQRFMRFLAHNGIFEIHESQEDHELTYALTPASKLLVNSSDHCLSPMVLAF 386
Query: 120 TNPTGVELYYQLTKWTSKEDLTIFDT----TLWDFMQQNSAYAELFNDAMESDSNMMRFA 175
T+P Y+ L +W ED ++F+T + W +++N Y LFN+AM SDS ++ A
Sbjct: 387 TDPLRNVKYHHLGEWIRGEDPSVFETAHGTSAWGLLEKNPEYFSLFNEAMASDSRIVDLA 566
Query: 176 MSDCKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGG 235
+ +C SVFEG+ S+VDVGGGTG T +II +AFP LKC+V DLP+VVE LTG N LSFVGG
Sbjct: 567 LKNCTSVFEGLDSMVDVGGGTGTTARIICDAFPKLKCVVLDLPHVVENLTGTNNLSFVGG 746
Query: 236 NMFESIPQADAILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEH 286
+MF SIPQADA+LLK +C+KIL+KC+++ S + GKVIIID VINEK D+
Sbjct: 747 DMFNSIPQADAVLLKWVLHNWTDENCIKILQKCRDSISSKGNSGKVIIIDAVINEKLDDP 926
Query: 287 EITEVKLCFDITMMANHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIEL 339
++T+ KL DI M+ N RER EK WK++ EAGF YKIFPIFGFRSLIE+
Sbjct: 927 DMTQTKLSLDIIML-TMNGRERTEKEWKQLFIEAGFKHYKIFPIFGFRSLIEV 1082
>TC210019 similar to UP|Q84KK5 (Q84KK5) S-adenosyl-L-methionine: daidzein
7-0-methyltransferase, complete
Length = 1156
Score = 363 bits (931), Expect = e-101
Identities = 190/350 (54%), Positives = 246/350 (70%), Gaps = 13/350 (3%)
Frame = +1
Query: 3 SENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSALRI 62
S N SE+F Q LY +++F+ + +K VELGIPDII NHG+PITLPELVS L+I
Sbjct: 25 SINGRNESEIFQGQTLLYKHLYAFIDSMCLKCIVELGIPDIIHNHGQPITLPELVSILQI 204
Query: 63 PEAKAGCVHSVMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNP 122
P AK V S+MR L HN F V+I + KEAYAL+ S+LLVK ++ L M++ V +P
Sbjct: 205 PPAKVSQVQSLMRYLEHNGFFERVRIHE-KEAYALTAASELLVKSSELSLAPMIEFVLDP 381
Query: 123 TGVELYYQLTKWTSKEDLTIFDTTL----WDFMQQNSAYAELFNDAMESDSNMMRFAMSD 178
T ++QL KW ++DLT+FD +L WDF+ +N A+ + FN+AM SDS MM A+ D
Sbjct: 382 TLSNSFHQLKKWVYEKDLTLFDISLGSHLWDFLNKNPAHNKSFNEAMASDSQMMNLALRD 561
Query: 179 CKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMF 238
C VF+G+ +VDVGGGTG T II EAFP LKCIVFD P V+E L+G+N L++VGG+MF
Sbjct: 562 CNWVFQGLEFIVDVGGGTGTTAXIICEAFPNLKCIVFDRPQVIENLSGSNNLTYVGGDMF 741
Query: 239 ESIPQADAILL---------KDCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEIT 289
+SIP+AD ILL KDC+KILK CKEA S K GKVIIID+VINEK+DEH++T
Sbjct: 742 KSIPKADVILLKWILHNWIDKDCIKILKNCKEAISNNGKRGKVIIIDVVINEKEDEHKVT 921
Query: 290 EVKLCFDITMMANHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIEL 339
E+KL DIT MA N +ER+E+ WKK+ EAGF YKIFP+ + S+IE+
Sbjct: 922 ELKLVMDIT-MACVNGKERNEEEWKKLFMEAGFQDYKIFPLTKYLSVIEI 1068
>TC206764 similar to UP|7MT9_MEDSA (O22309) Isoflavone-7-O-methytransferase 9
(Isoflavone-O-methytransferase 9) (7 IOMT-9) , partial
(86%)
Length = 1210
Score = 347 bits (890), Expect = 4e-96
Identities = 182/350 (52%), Positives = 235/350 (67%), Gaps = 15/350 (4%)
Frame = +1
Query: 5 NEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNH--GKPITLPELVSALRI 62
N KASE+F QA LY + F+ +KW VEL IPDII +H G+PIT ELVS L++
Sbjct: 58 NGRKASEIFQGQALLYKHLLGFIDSKCLKWMVELDIPDIIHSHSHGQPITFSELVSILQV 237
Query: 63 PEAKAGCVHSVMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNP 122
P K V S+MR LAHN F IV+I DN EAYAL+ S+LLVK ++ L MV+ P
Sbjct: 238 PPTKTRQVQSLMRYLAHNGFFEIVRIHDNIEAYALTAASELLVKSSELSLAPMVEYFLEP 417
Query: 123 TGVELYYQLTKWTSKEDLTIFDTTL----WDFMQQNSAYAELFNDAMESDSNMMRFAMSD 178
+ QL +W +EDLT+F+ +L WDF+ ++ AY + FN+AM DS M+ A D
Sbjct: 418 NCQGAWNQLKRWVHEEDLTVFEVSLGTPFWDFINKDPAYNKSFNEAMACDSQMLNLAFRD 597
Query: 179 CKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMF 238
C VFEG+ S+VDVGGGTG T KII EAFP LKC+V + PNVVE L+G+N L+FVGG+MF
Sbjct: 598 CNWVFEGLESIVDVGGGTGITAKIICEAFPKLKCMVLERPNVVENLSGSNNLTFVGGDMF 777
Query: 239 ESIPQADAILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEIT 289
+ IP+ADA+LLK DC+KIL+ CKEA S + K GKV++ID VINE +DE ++T
Sbjct: 778 KCIPKADAVLLKLVLHNWNDNDCMKILENCKEAISGESKTGKVVVIDTVINENKDERQVT 957
Query: 290 EVKLCFDITMMANHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIEL 339
E+KL D+ M N +ER E+ WKK+ EAGF SYKI P G+ SLIE+
Sbjct: 958 ELKLLMDVHMACIINGKERKEEDWKKLFMEAGFQSYKISPFTGYLSLIEI 1107
>TC216576 similar to UP|Q84KK6 (Q84KK6) S-adenosyl-L-methionine:
2,7,4'-trihydroxyisoflavanone 4'-O-methyltransferase,
complete
Length = 1354
Score = 311 bits (797), Expect = 3e-85
Identities = 167/358 (46%), Positives = 238/358 (65%), Gaps = 23/358 (6%)
Frame = +1
Query: 5 NEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSALRIPE 64
N + SEL AQ HLY +++F+ +++K A+ELGI D+I +HGKP+T+ EL SAL++
Sbjct: 67 NSTEESELHHAQIHLYKHVYNFVSSMALKSAMELGIADVIHSHGKPMTISELSSALKLHP 246
Query: 65 AKAGCVHSVMRLLAHNKIFAIV------KIDDNKE-AYALSPTSKLLVKGTDHCLTSMVK 117
+K + +RLL HN FA ++ +E AYAL+P SKLL++ CL +VK
Sbjct: 247 SKVSVLQRFLRLLTHNGFFAKTILPSKNGVEGGEEIAYALTPPSKLLIRNKSICLAPIVK 426
Query: 118 MVTNPTGVELYYQLTKWTSKE-DLTIFDT----TLWDFMQQNSAYAEL--FNDAMESDSN 170
+ + +++++ KW S++ +LT++++ + WDF+ + + L F DAM +DS
Sbjct: 427 GALHSSSLDMWHSSKKWFSEDKELTLYESATGESFWDFLNKTTESDTLGMFQDAMAADSK 606
Query: 171 MMRFAMSDCKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYL 230
+ + A+ +CK VFEG+ SLVDVGGGTG ++ISE FP LKC VFD P VV LTGN L
Sbjct: 607 VFKLALEECKHVFEGLGSLVDVGGGTGVVTRLISETFPHLKCTVFDQPQVVANLTGNENL 786
Query: 231 SFVGGNMFESIPQADAILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINE 281
+FVGG+MF+SIP ADA+LLK VKILK CKEA S + K GKVIIIDI I+E
Sbjct: 787 NFVGGDMFKSIPSADAVLLKWVLHDWNDELSVKILKNCKEAISGKGKEGKVIIIDIAIDE 966
Query: 282 KQDEHEITEVKLCFDITMMANHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIEL 339
D+ E+TE+KL +D+ M+ N +ER++K W+K+I EAGF +YKI PI GF+SLIE+
Sbjct: 967 VGDDREMTELKLDYDLVMLTMFNGKEREKKEWEKLIYEAGFSNYKIIPICGFKSLIEV 1140
>TC205647 similar to UP|Q8GU23 (Q8GU23) Orcinol O-methyltransferase, partial
(89%)
Length = 1774
Score = 285 bits (730), Expect = 2e-77
Identities = 156/356 (43%), Positives = 221/356 (61%), Gaps = 17/356 (4%)
Frame = +2
Query: 1 MDSENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSAL 60
M+S +E A++L AQ H++N IFSF+ +S+K V+LGIPDII N+G+P+ L L+++L
Sbjct: 269 MESHDEEHAAKLLRAQTHIWNHIFSFINSMSLKCVVDLGIPDIIHNYGQPMPLSNLIASL 448
Query: 61 RIPEAKAGCVHSVMRLLAHNKIFAIVKIDDNKEA---YALSPTSKLLVKGTDHCLTSMVK 117
I +K VH +MR++ H+ F+ K D E Y L+ S LL+K +T +
Sbjct: 449 PIHPSKTCFVHRLMRIMIHSGFFSQQKHDLENELEAKYVLTDASVLLLKNHPMSVTPFLH 628
Query: 118 MVTNPTGVELYYQLTKWTSKEDLTIFDTT----LWDFMQQNSAYAELFNDAMESDSNMMR 173
+ +P + Q + W D T F+T LWD+ + LFNDAM SD+ +
Sbjct: 629 AMLDPVLTNPWNQFSTWFKNGDPTPFETAHGMMLWDYAGADPKLNNLFNDAMASDARFVT 808
Query: 174 -FAMSDCKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSF 232
+ CK VF G+ SLVDVGGGTG K I+++FP ++CIVFDLP+VV GL G+ L +
Sbjct: 809 SLVIEKCKGVFMGLESLVDVGGGTGTMAKAIAKSFPRVECIVFDLPHVVSGLKGSENLKY 988
Query: 233 VGGNMFESIPQADAILL---------KDCVKILKKCKEAASRQKKGGKVIIIDIVINEKQ 283
V G+MFE+IP ADAILL K+CV ILKKCKEA +R+ K GKVIIID+V+ ++
Sbjct: 989 VAGDMFEAIPPADAILLKWILHDWNDKECVDILKKCKEAITRKGKEGKVIIIDMVVENEK 1168
Query: 284 DEHEITEVKLCFDITMMANHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIEL 339
+ E E +L FD+ MM +ER +K W K+I+ AG+ +YKI P+ G RSLIE+
Sbjct: 1169RDDESVETQLFFDMLMMVLVTGKERSKKEWAKLISSAGYNNYKITPVLGLRSLIEI 1336
>TC206765 similar to UP|Q84KK5 (Q84KK5) S-adenosyl-L-methionine: daidzein
7-0-methyltransferase, partial (54%)
Length = 584
Score = 187 bits (474), Expect = 8e-48
Identities = 100/195 (51%), Positives = 130/195 (66%), Gaps = 4/195 (2%)
Frame = +3
Query: 12 LFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSALRIPEAKAGCVH 71
+F Q LY +F+ + +K VELGIPDII NHG+ ITLPEL S L+IP AK V
Sbjct: 3 IFQGQTLLYKHLFAHVDSKCLKCIVELGIPDIIHNHGQAITLPELASILQIPPAKVSQVQ 182
Query: 72 SVMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNPTGVELYYQL 131
S+MR LAHN F V I + KEAYAL+ S+LLVK ++ L MV+ + + T ++QL
Sbjct: 183 SLMRYLAHNGFFERVTIHE-KEAYALTAASELLVKSSELSLAPMVEYILDTTISGSFHQL 359
Query: 132 TKWTSKEDLTIFDTTL----WDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEGIT 187
KW +EDLT+F+ +L WDF+ +N AY + FN+AM SDS M+ A+ DCK VFEG+
Sbjct: 360 KKWVHEEDLTLFEISLGSHLWDFLNRNPAYNKSFNEAMASDSQMLNLALRDCKLVFEGLE 539
Query: 188 SLVDVGGGTGNTVKI 202
S+VDVGGGTG T KI
Sbjct: 540 SIVDVGGGTGATAKI 584
>TC210492 weakly similar to UP|Q8GU23 (Q8GU23) Orcinol O-methyltransferase,
partial (51%)
Length = 710
Score = 172 bits (435), Expect = 3e-43
Identities = 91/207 (43%), Positives = 133/207 (63%), Gaps = 13/207 (6%)
Frame = +1
Query: 146 TLWDFMQQNSAYAELFNDAMESDSNMMRFAMS----DCKSVFEGITSLVDVGGGTGNTVK 201
T +D+ +++ + +NDAM D+ RFA S + K VFEG+ S+VDVGGG G K
Sbjct: 10 TYFDYAKRDPKFGHFYNDAMAKDT---RFASSVVIENYKEVFEGLKSIVDVGGGIGTMAK 180
Query: 202 IISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILLK---------DC 252
I++AFP +KC VFDLP+VV+GL G + +VGG+MFE IP AD I+LK +C
Sbjct: 181 AIAKAFPQVKCTVFDLPHVVDGLQGTENIEYVGGDMFEVIPAADCIMLKWVLHCWNDEEC 360
Query: 253 VKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEITEVKLCFDITMMANHNSRERDEKT 312
+KILKKCKEA GKVII+++V+ ++++++ E++LC D+ MM+ ++R EK
Sbjct: 361 MKILKKCKEAI---PSDGKVIIMELVMEHNKEDNKLIEMQLCCDMLMMSLFAGKDRTEKE 531
Query: 313 WKKIITEAGFMSYKIFPIFGFRSLIEL 339
W +I AGF +YKI IF +IE+
Sbjct: 532 WAHLIASAGFSNYKITHIFDLYHIIEV 612
>TC203610 weakly similar to UP|Q7X9I9 (Q7X9I9) Caffeic acid
O-methyltransferase, partial (62%)
Length = 1504
Score = 159 bits (402), Expect = 2e-39
Identities = 107/343 (31%), Positives = 179/343 (51%), Gaps = 18/343 (5%)
Frame = +1
Query: 14 VAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSALR-IPEAKAGCVHS 72
+ Q ++ + F V++K +EL I DI+ +GKP++L ++V + P A +
Sbjct: 202 LGQVEIWRYMTCFTDSVALKAVIELRIADILDRYGKPLSLSQIVENIEDAPSPDASLLQR 381
Query: 73 VMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNPTGVELYY--- 129
V+R++ KIF+ + + + + L+ SK +++ T L M+ + +P + +
Sbjct: 382 VLRVMVRRKIFSAQESETGETLFGLTRASKWILRDTKMTLAPMLLLENHPIHLNPAHYIS 561
Query: 130 QLTKWTSKEDLTIFDT---TLWDFMQQNSAYAELFNDAMESDSNMMRFAM-SDCKSVFEG 185
++ + +K F ++ + Y LFN+ M + ++ A+ + K F
Sbjct: 562 EIIREGTKNGTAFFKCHGHEQFEMTGLDPEYNRLFNEGMVCTARVVSKAVITGYKDGFNQ 741
Query: 186 ITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQAD 245
I SLVDVGGG G ++ I A+P + I FDLP+VV + ++ VGG+MF SIP AD
Sbjct: 742 IKSLVDVGGGIGGSLSEIVRAYPHINAINFDLPHVVATAPKFDGITHVGGDMFVSIPSAD 921
Query: 246 AILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEITEVKLCFD 296
AI +K C+KILK C++A +K GKVII+D V+ + +E T+V + FD
Sbjct: 922 AIYMKWILHDWSDEHCIKILKNCRKAI--PEKTGKVIIVDHVLRPEGNE-LFTDVGIAFD 1092
Query: 297 ITMMA-NHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIE 338
+ ++A N +ER E+ WK + E GF Y I I S+IE
Sbjct: 1093MMLLAHNAGGKERTEENWKWLFKETGFARYNIIKINALPSIIE 1221
>TC213916 similar to UP|Q8GU23 (Q8GU23) Orcinol O-methyltransferase, partial
(46%)
Length = 738
Score = 159 bits (401), Expect = 2e-39
Identities = 86/170 (50%), Positives = 105/170 (61%), Gaps = 9/170 (5%)
Frame = +1
Query: 179 CKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMF 238
CK VF G SLVDVGGGTG + FP L+C VFDLP+VV L G+ L +VGG+M
Sbjct: 13 CKGVFNGWESLVDVGGGTGTXXMAMPXXFPQLECTVFDLPHVVATLQGSENLKYVGGDMX 192
Query: 239 ESIPQADAILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEIT 289
ESIP ADAILLK CVKILKKCKEA KVIIID+V+ ++ + E
Sbjct: 193 ESIPSADAILLKWILHDWNDEQCVKILKKCKEAIK-----SKVIIIDMVVENEKGDDESI 357
Query: 290 EVKLCFDITMMANHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIEL 339
E +L D+ +M + +ER EK W K+I GF YKI P+ G RSLIE+
Sbjct: 358 ETQLFIDMVVMVLYPGKERTEKEWAKLIFSTGFSDYKITPVLGLRSLIEI 507
>AW733746 similar to GP|28804594|db S-adenosyl-L-methionine: daidzein
7-0-methyltransferase {Glycyrrhiza echinata}, partial
(37%)
Length = 411
Score = 154 bits (388), Expect = 7e-38
Identities = 74/128 (57%), Positives = 96/128 (74%), Gaps = 9/128 (7%)
Frame = +2
Query: 146 TLWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEGITSLVDVGGGTGNTVKIISE 205
+LWDF+ +N AY E FN+AM DS M A+ DCK VFEG+ S+VDVGGGTG T ++ISE
Sbjct: 23 SLWDFLNKNPAYNESFNEAMARDSQMSNLALRDCKLVFEGLESIVDVGGGTGATARMISE 202
Query: 206 AFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILL---------KDCVKIL 256
AFP LKC+V D P+V+E L+ +N L++VGG+MF+SIP+ADA+LL KDC+KIL
Sbjct: 203 AFPDLKCVVLDRPHVLENLSESNNLTYVGGDMFKSIPKADAVLLKWILHDWTDKDCIKIL 382
Query: 257 KKCKEAAS 264
+ CKEA S
Sbjct: 383 ENCKEAIS 406
>TC221589 weakly similar to UP|O65859 (O65859) O-methyltransferase, partial
(52%)
Length = 693
Score = 142 bits (359), Expect = 2e-34
Identities = 79/227 (34%), Positives = 130/227 (56%), Gaps = 10/227 (4%)
Frame = +1
Query: 4 ENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSALRIP 63
+NE ++L AQ H+++Q F F+ +S+K A++L IPD+I +G+P+ L +L+++L I
Sbjct: 13 DNEDHYAKLLRAQTHIFDQTFGFINSMSLKCAIDLCIPDVIHKYGQPMPLSQLIASLPIH 192
Query: 64 EAKAGCVHSVMRLLAHNKIF-----AIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKM 118
+KA + +M++L H+ F A + + +Y L+ SKLL+K + S+ ++
Sbjct: 193 PSKACFISRLMQILTHSGFFSQHNNATENYEQEEVSYVLTDASKLLLKDHHFSMISLPQV 372
Query: 119 VTNPTGVELYYQLTKWTSKEDLTIFDT----TLWDFMQQNSAYAELFNDAMESDSNMMRF 174
+ +P V ++Q + W + ED T F T WD+ LFNDAM +DS ++
Sbjct: 373 ILDPILVNPWFQFSTWFTNEDPTPFHTQNGMAFWDYASSEPKLNHLFNDAMTNDSRLISS 552
Query: 175 AMSD-CKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNV 220
+ + CK VF G+ SLVDVGGGTG+ I+ F LKC V L ++
Sbjct: 553 VLIEKCKGVFSGLESLVDVGGGTGSLALAIA*TFA*LKCFVCVLVHI 693
>BG509170 similar to GP|28804594|db S-adenosyl-L-methionine: daidzein
7-0-methyltransferase {Glycyrrhiza echinata}, partial
(36%)
Length = 412
Score = 142 bits (358), Expect = 2e-34
Identities = 72/129 (55%), Positives = 93/129 (71%), Gaps = 9/129 (6%)
Frame = +2
Query: 220 VVEGLTGNNYLSFVGGNMFESIPQADAILLK---------DCVKILKKCKEAASRQKKGG 270
VVE L+G+N L++VGG+MF+SIP+A A+L K DC KIL+ CKEA S + K G
Sbjct: 2 VVENLSGSNNLTYVGGDMFKSIPKACAVLFKMILHNWSDEDCRKILENCKEAISSKSKTG 181
Query: 271 KVIIIDIVINEKQDEHEITEVKLCFDITMMANHNSRERDEKTWKKIITEAGFMSYKIFPI 330
KVI+ID+VINEK+DEHEIT +KL D+ M N +ER E+ WKK+ EAGF SYKI P+
Sbjct: 182 KVIVIDVVINEKKDEHEITRLKLLMDLNMACLLNGKERREEDWKKLFVEAGFQSYKISPL 361
Query: 331 FGFRSLIEL 339
G+ SLIE+
Sbjct: 362 TGYLSLIEI 388
>BM528698 similar to GP|28804594|db S-adenosyl-L-methionine: daidzein
7-0-methyltransferase {Glycyrrhiza echinata}, partial
(40%)
Length = 432
Score = 137 bits (345), Expect = 7e-33
Identities = 69/140 (49%), Positives = 98/140 (69%)
Frame = +3
Query: 8 KASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSALRIPEAKA 67
KA+E+F QA LY +F+F+ + +K +ELGIPDII HG+PITL ELVS L +P A+
Sbjct: 9 KATEIFQGQAILYRCMFAFLDSMCLKSIIELGIPDIIHKHGQPITLSELVSILNVPPARV 188
Query: 68 GCVHSVMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNPTGVEL 127
G V S+MR LAH+ F ++I KE+YAL+ S+LLVK ++ CLT MV+ V +PT
Sbjct: 189 GHVQSLMRYLAHHGFFERLRIHLEKESYALTAASELLVKSSELCLTPMVEKVLDPTLSAS 368
Query: 128 YYQLTKWTSKEDLTIFDTTL 147
++Q+ KW +EDL++FD +L
Sbjct: 369 FHQMKKWVYEEDLSVFDISL 428
>CA953078 weakly similar to GP|28804592|dbj S-adenosyl-L-methionine: 2 7
4'-trihydroxyisoflavanone 4'-O-methyltransferase
{Glycyrrhiza echinata}, partial (12%)
Length = 427
Score = 129 bits (325), Expect = 1e-30
Identities = 72/133 (54%), Positives = 90/133 (67%), Gaps = 2/133 (1%)
Frame = +3
Query: 1 MDSENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSAL 60
MD++N A E F Q LY QIF +RPV + WA ELGIPDII NHGKPITL ELVSAL
Sbjct: 33 MDNQN---AIEFFEGQNLLYMQIFGNLRPVCLMWACELGIPDIISNHGKPITLLELVSAL 203
Query: 61 RIPEAKAGCVHSVMRLLAHNKIFAI--VKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKM 118
+IP +K G V MR LAHN+IF I + D ++ AYAL+P SKLLV + HCL+ M++
Sbjct: 204 QIPPSKVGFVKRFMRFLAHNRIFDIHESQEDHHELAYALTPASKLLVNDSIHCLSPMLQF 383
Query: 119 VTNPTGVELYYQL 131
+T+P Y+ L
Sbjct: 384 MTDPFLTNAYHHL 422
>TC226262 similar to PRF|2119166A.0|1582580|2119166A caffeic acid
O-methyltransferase. {Stylosanthes humilis;} , complete
Length = 1331
Score = 123 bits (308), Expect = 1e-28
Identities = 92/320 (28%), Positives = 157/320 (48%), Gaps = 21/320 (6%)
Frame = +2
Query: 29 PVSIKWAVELGIPDIIQNHGKPITLPELVSALRIPEAKAGC---VHSVMRLLAHNKI--F 83
P+ +K A+EL + +II G + L A R+P + ++RLLA I F
Sbjct: 122 PMVLKSALELDLLEIIAKAGPGVHLSPSDIASRLPTHNPDAPVMLDRILRLLACYNILSF 301
Query: 84 AIVKIDDNK--EAYALSPTSKLLVKGTDHCLTSMVKMVT-NPTGVELYYQLTKWTSKEDL 140
++ + K Y L+P +K LV+ D + + ++ + +E +Y L + +
Sbjct: 302 SLRTLPHGKVERLYGLAPVAKYLVRNEDGVSIAALNLMNQDKILMESWYYLKDAVLEGGI 481
Query: 141 TI---FDTTLWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEGITSLVDVGGGTG 197
+ T +++ + + ++FN M S + + + + FE + SLVDVGGGTG
Sbjct: 482 PFNKAYGMTAFEYHGTDPRFNKVFNKGMADHSTITMKKILETYTGFESLKSLVDVGGGTG 661
Query: 198 NTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILLK------- 250
+ +I PT+K I FDLP+V+E + VGG+MF S+P+ADAI +K
Sbjct: 662 AVINMIVSKHPTIKGINFDLPHVIEDAPSYPGVEHVGGDMFASVPKADAIFMKWICHDWS 841
Query: 251 --DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEITEVKLCFDITMMA-NHNSRE 307
C+K LK C EA GKVI+ + ++ D T+ + D+ M+A N +E
Sbjct: 842 DEHCLKFLKNCYEAL---PDNGKVIVAECILPVAPDSSLATKGVVHIDVIMLAHNPGGKE 1012
Query: 308 RDEKTWKKIITEAGFMSYKI 327
R EK ++ + +GF +++
Sbjct: 1013RTEKEFEALAKGSGFQGFRV 1072
>TC207440 weakly similar to UP|Q8GU21 (Q8GU21) Orcinol O-methyltransferase
(Fragment), partial (26%)
Length = 938
Score = 122 bits (307), Expect = 2e-28
Identities = 73/222 (32%), Positives = 122/222 (54%), Gaps = 15/222 (6%)
Frame = +3
Query: 73 VMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNPTGVELYYQLT 132
+MR L KIF K D Y+ S S+ L++ +H + S + + ++P + ++ L+
Sbjct: 54 IMRFLVQRKIF---KGDGCSRGYSQSALSRRLMRNGEHSMASFLLLESSPVMLAPWHSLS 224
Query: 133 KWTSKEDLTIFDTT----LWDFMQQNSAYAELFNDAMESDSNM-MRFAMSDCKSVFEGIT 187
F +W + N ++ L N+AM D+ + M + C F G+
Sbjct: 225 ARVMANGNPSFAKAHGEDVWRYAAANLDHSNLINEAMACDAKLVMPIIIQSCSEAFHGLK 404
Query: 188 SLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGN-NYLSFVGGNMFESIPQADA 246
SLVDVGGG G ++I+++A P+++ I FDLP+V+ G+ + + V G+MF S+P+ADA
Sbjct: 405 SLVDVGGGNGTAMRILAKACPSIRPINFDLPHVIALCDGDGDGVQHVSGDMFLSVPKADA 584
Query: 247 ILL---------KDCVKILKKCKEAASRQKKGGKVIIIDIVI 279
L ++C++ILKKC+EA S K+ G+VII++ VI
Sbjct: 585 AFLMWVLHDWSDEECIQILKKCREAISNSKENGRVIIVEAVI 710
>BU091459 weakly similar to GP|28804594|db S-adenosyl-L-methionine: daidzein
7-0-methyltransferase {Glycyrrhiza echinata}, partial
(39%)
Length = 430
Score = 119 bits (297), Expect = 3e-27
Identities = 63/143 (44%), Positives = 94/143 (65%), Gaps = 4/143 (2%)
Frame = +1
Query: 23 IFSFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSALRIPEAKAGCVHSVMRLLAHNKI 82
+F+F+ + +K +ELGIPDII HG+PITL ELVS L +P A+ G V S+M L+H++
Sbjct: 4 MFAFIDSMCLKTIIELGIPDIIHKHGQPITLSELVSILHVPPARVGHVQSLMHYLSHHRF 183
Query: 83 FAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNPTGVELYYQLTKWTSKEDLTI 142
F V+I + KEAYAL+ S+LLVK ++ L MVK + +PT ++Q+ KW ++DL++
Sbjct: 184 FESVRIHE-KEAYALTAASELLVKSSELSLAPMVKYILDPTLSASFHQMKKWVYEKDLSV 360
Query: 143 FD----TTLWDFMQQNSAYAELF 161
FD +L + +N AY E F
Sbjct: 361 FDISLGCSLMVXLNKNPAYNESF 429
>TC203345 weakly similar to UP|Q7X9I9 (Q7X9I9) Caffeic acid
O-methyltransferase, partial (48%)
Length = 784
Score = 116 bits (290), Expect = 2e-26
Identities = 72/169 (42%), Positives = 98/169 (57%), Gaps = 10/169 (5%)
Frame = +1
Query: 180 KSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFE 239
K F I SLVDVGGG G ++ I A+P + I FDLP+VV + ++ VGG+MF
Sbjct: 34 KDGFNQIKSLVDVGGGIGGSLSEIVRAYPHINAINFDLPHVVATAPKYDGITHVGGDMFV 213
Query: 240 SIPQADAILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEITE 290
SIP ADAI +K CVKILK C++A +K GKVII+D V+ + +E T+
Sbjct: 214 SIPDADAIYMKWILHDWSDEHCVKILKNCRKAI--PEKTGKVIIVDHVLRPEGNE-LFTD 384
Query: 291 VKLCFDITMMA-NHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIE 338
V + FD+ ++A N +ER E+ WK + E GF Y I I S+IE
Sbjct: 385 VGIAFDMMLLAHNAGGKERTEENWKWLFKETGFARYNIIKINALPSIIE 531
>TC216384 weakly similar to PRF|1906376A.0|444327|1906376A
O-methyltransferase. {Populus balsamifera subsp.
trichocarpa x Populus deltoides;} , partial (40%)
Length = 1422
Score = 114 bits (284), Expect = 8e-26
Identities = 91/335 (27%), Positives = 151/335 (44%), Gaps = 31/335 (9%)
Frame = +3
Query: 24 FSFMRPVSIKWAVELGIPDIIQN-HGKPITLPELVSALRIPEAK-AGCVHSVMRLLA--- 78
FS + P + AV+L + DII ++ E+ S L P + A + ++ +LA
Sbjct: 135 FSRIFPAILNAAVDLNLFDIIDKAESSTLSASEIASLLPNPHPQLANRLERILPVLASYS 314
Query: 79 -------------HNKIFAIVKIDD----NKEAYALSPTSKLLVKGTDHCLTSMVKMVTN 121
+++A+ I + + +L P S L +G H L + + +
Sbjct: 315 LLNCSIRTTEDGVRERLYALSPIGQYFASDDDGGSLGPLSSLFHRGYFHVLKDVKDAIMD 494
Query: 122 PTGVELYYQLTKWTSKEDLTIFDTTLWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKS 181
P + + + + +M+ + +LFN A+ +
Sbjct: 495 PNNNDHFQN-----------VHGMPPYQYMKTDEELNKLFNKALAQTGPPAMKMLLKLYK 641
Query: 182 VFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESI 241
FE +++LVDVGGG G T+K I +P++K I FDLP VV+ + V G+MFES+
Sbjct: 642 GFEQVSTLVDVGGGVGETLKQIIFEYPSIKGINFDLPQVVQDAPPYPGIEHVEGDMFESV 821
Query: 242 PQADAILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEITEVK 292
P+ DAILLK DCVK L+ C +A + GKVI+ID +I E D +I+
Sbjct: 822 PKGDAILLKLVCHNWLDEDCVKFLRNCHKALPQH---GKVIVIDYIIPEVPDSSKISMQT 992
Query: 293 LCFDITMMANHNSRERDEKTWKKIITEAGFMSYKI 327
D M + +ER EK ++ + +GF + +
Sbjct: 993 CVADSLMFLVTSGKERTEKEFESLCRNSGFSRFHV 1097
>BQ298504
Length = 420
Score = 107 bits (268), Expect = 6e-24
Identities = 52/90 (57%), Positives = 68/90 (74%)
Frame = +2
Query: 250 KDCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEITEVKLCFDITMMANHNSRERD 309
++C+KIL+KC+++ S + GKVIIID VINEK D+ ++T+ KL DI M+ N RER
Sbjct: 149 ENCIKILQKCRDSISSKGNSGKVIIIDAVINEKLDDPDMTQTKLSLDIIMLTM-NGRERT 325
Query: 310 EKTWKKIITEAGFMSYKIFPIFGFRSLIEL 339
EK WK++ EAGF YKIFPIFGFRSLIE+
Sbjct: 326 EKEWKQLFIEAGFKHYKIFPIFGFRSLIEV 415
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,583,985
Number of Sequences: 63676
Number of extensions: 163471
Number of successful extensions: 753
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 707
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 708
length of query: 341
length of database: 12,639,632
effective HSP length: 98
effective length of query: 243
effective length of database: 6,399,384
effective search space: 1555050312
effective search space used: 1555050312
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144475.7


