

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
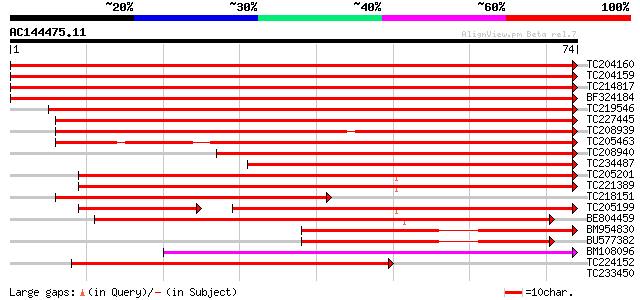
Query= AC144475.11 - phase: 0 /pseudo
(74 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 153 1e-38
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 151 7e-38
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 149 2e-37
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 149 2e-37
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 133 1e-32
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 107 9e-25
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 99 5e-22
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 93 3e-20
TC208940 UP|Q39803 (Q39803) BiP isoform C (Fragment), partial (26%) 79 3e-16
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 71 9e-14
TC205201 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa prote... 70 3e-13
TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa p... 68 1e-12
TC218151 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 62 7e-11
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 49 2e-09
BE804459 similar to PIR|T10248|T10 heat shock protein 70K chlo... 53 3e-08
BM954830 similar to GP|15620767|emb heat shock protein (Hsp70) {... 47 2e-06
BU577382 47 2e-06
BM108096 43 3e-05
TC224152 similar to UP|Q8L8N8 (Q8L8N8) 70kD heat shock protein, ... 42 6e-05
TC233450 38 0.001
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 153 bits (387), Expect = 1e-38
Identities = 73/74 (98%), Positives = 74/74 (99%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 95 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 274
Query: 61 QVAMNPTNTVFDAK 74
QVAMNPTNTVFDAK
Sbjct: 275 QVAMNPTNTVFDAK 316
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 151 bits (381), Expect = 7e-38
Identities = 72/74 (97%), Positives = 73/74 (98%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 80 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 259
Query: 61 QVAMNPTNTVFDAK 74
QVAMNP NTVFDAK
Sbjct: 260 QVAMNPINTVFDAK 301
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 149 bits (377), Expect = 2e-37
Identities = 71/74 (95%), Positives = 72/74 (96%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYV FTD+ERLIGDAAKN
Sbjct: 86 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKN 265
Query: 61 QVAMNPTNTVFDAK 74
QVAMNP NTVFDAK
Sbjct: 266 QVAMNPINTVFDAK 307
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 149 bits (377), Expect = 2e-37
Identities = 71/74 (95%), Positives = 72/74 (96%)
Frame = -3
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYV FTD+ERLIGDAAKN
Sbjct: 369 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKN 190
Query: 61 QVAMNPTNTVFDAK 74
QVAMNP NTVFDAK
Sbjct: 189 QVAMNPINTVFDAK 148
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 133 bits (335), Expect = 1e-32
Identities = 64/69 (92%), Positives = 66/69 (94%)
Frame = +1
Query: 6 EGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMN 65
EG AIGIDLGTTYSCVGVWQ+DRVEII NDQGNRTTPSYVAFTD+ERLIGDAAKNQVAMN
Sbjct: 13 EGKAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVAMN 192
Query: 66 PTNTVFDAK 74
P NTVFDAK
Sbjct: 193PQNTVFDAK 219
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 107 bits (268), Expect = 9e-25
Identities = 51/68 (75%), Positives = 58/68 (85%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+VAFTDSERLIG+AAKN A+NP
Sbjct: 209 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLAAVNP 388
Query: 67 TNTVFDAK 74
T+FD K
Sbjct: 389 ERTIFDVK 412
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 98.6 bits (244), Expect = 5e-22
Identities = 48/68 (70%), Positives = 56/68 (81%)
Frame = +3
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+ +FTDSERLIG+AAKN A+NP
Sbjct: 171 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSW-SFTDSERLIGEAAKNLAAVNP 347
Query: 67 TNTVFDAK 74
+FD K
Sbjct: 348 ERVIFDVK 371
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 92.8 bits (229), Expect = 3e-20
Identities = 50/68 (73%), Positives = 54/68 (78%)
Frame = +3
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDL TTYSCVGV D VEIIANDQGNR TPS+VAFTDSERLIG+AAK A+NP
Sbjct: 192 GTVIGIDL-TTYSCVGVT--DAVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVAAVNP 362
Query: 67 TNTVFDAK 74
T+FD K
Sbjct: 363 VRTIFDVK 386
>TC208940 UP|Q39803 (Q39803) BiP isoform C (Fragment), partial (26%)
Length = 447
Score = 79.3 bits (194), Expect = 3e-16
Identities = 37/47 (78%), Positives = 42/47 (88%)
Frame = +1
Query: 28 RVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNTVFDAK 74
+VEIIANDQGNR TPS+VAFTDSERLIG+AAKNQ A+NP T+FD K
Sbjct: 82 QVEIIANDQGNRITPSWVAFTDSERLIGEAAKNQAAVNPERTIFDVK 222
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 71.2 bits (173), Expect = 9e-14
Identities = 34/43 (79%), Positives = 36/43 (83%)
Frame = +1
Query: 32 IANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNTVFDAK 74
I NDQGN TTPS VAFTD +RLIG+AAKNQ A NP NTVFDAK
Sbjct: 1 IHNDQGNNTTPSCVAFTDQQRLIGEAAKNQAATNPENTVFDAK 129
>TC205201 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (18%)
Length = 467
Score = 69.7 bits (169), Expect = 3e-13
Identities = 36/66 (54%), Positives = 43/66 (64%), Gaps = 1/66 (1%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD-SERLIGDAAKNQVAMNPTN 68
IGIDLGTT SCV V + ++I N +G RTTPS VAF +E L+G AK Q NPTN
Sbjct: 247 IGIDLGTTNSCVSVMEGKNPKVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTN 426
Query: 69 TVFDAK 74
T+F K
Sbjct: 427 TLFGTK 444
>TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (36%)
Length = 827
Score = 67.8 bits (164), Expect = 1e-12
Identities = 35/66 (53%), Positives = 41/66 (62%), Gaps = 1/66 (1%)
Frame = +3
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD-SERLIGDAAKNQVAMNPTN 68
IGIDLGTT SCV V + ++I N +G RTTPS VAF E L+G K Q NPTN
Sbjct: 255 IGIDLGTTNSCVSVMEGKNPKVIENSEGARTTPSVVAFNQKGELLVGTPXKRQAVTNPTN 434
Query: 69 TVFDAK 74
T+F K
Sbjct: 435 TLFGTK 452
>TC218151 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, partial (35%)
Length = 922
Score = 61.6 bits (148), Expect = 7e-11
Identities = 28/36 (77%), Positives = 31/36 (85%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTP 42
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TP
Sbjct: 190 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITP 297
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 48.5 bits (114), Expect(2) = 2e-09
Identities = 24/46 (52%), Positives = 30/46 (65%), Gaps = 1/46 (2%)
Frame = +3
Query: 30 EIIANDQGNRTTPSYVAFTD-SERLIGDAAKNQVAMNPTNTVFDAK 74
++I N +G RTTPS VAF +E L+G AK Q NPTNT+F K
Sbjct: 186 KVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTK 323
Score = 27.7 bits (60), Expect(2) = 2e-09
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQ 25
IGIDLGTT SCV V +
Sbjct: 49 IGIDLGTTNSCVSVME 96
>BE804459 similar to PIR|T10248|T10 heat shock protein 70K chloroplast -
cucumber, partial (18%)
Length = 478
Score = 53.1 bits (126), Expect = 3e-08
Identities = 28/61 (45%), Positives = 40/61 (64%), Gaps = 1/61 (1%)
Frame = +2
Query: 12 IDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPTNTV 70
IDLGTT V + + II+N +G +TTP A+T++ +RL+G AK QV +NP NT+
Sbjct: 116 IDLGTTNCSVAAFVGGKPTIISNSRGQKTTPFVRAYTNNGDRLVGLIAKRQVVVNPENTL 295
Query: 71 F 71
F
Sbjct: 296 F 298
>BM954830 similar to GP|15620767|emb heat shock protein (Hsp70)
{Moneuplotes crassus}, partial (3%)
Length = 406
Score = 47.0 bits (110), Expect = 2e-06
Identities = 26/36 (72%), Positives = 26/36 (72%)
Frame = +1
Query: 39 RTTPSYVAFTDSERLIGDAAKNQVAMNPTNTVFDAK 74
R TPSYVAFTDSE LIGD VAMNPTN VF K
Sbjct: 10 RPTPSYVAFTDSEPLIGD-----VAMNPTNIVFVRK 102
>BU577382
Length = 433
Score = 46.6 bits (109), Expect = 2e-06
Identities = 25/33 (75%), Positives = 25/33 (75%)
Frame = +1
Query: 39 RTTPSYVAFTDSERLIGDAAKNQVAMNPTNTVF 71
R TPSYVAFTDSE LIGD VAMNPTN VF
Sbjct: 1 RPTPSYVAFTDSEPLIGD-----VAMNPTNIVF 84
>BM108096
Length = 525
Score = 42.7 bits (99), Expect = 3e-05
Identities = 18/54 (33%), Positives = 29/54 (53%)
Frame = +3
Query: 21 VGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNTVFDAK 74
V V + ++++ ND+ R TP+ V F D +R +G A MNP N++ K
Sbjct: 9 VAVARQRGIDVVLNDESKRETPAIVCFGDKQRFLGTAGAASTMMNPKNSISQIK 170
>TC224152 similar to UP|Q8L8N8 (Q8L8N8) 70kD heat shock protein, partial
(33%)
Length = 563
Score = 42.0 bits (97), Expect = 6e-05
Identities = 18/42 (42%), Positives = 26/42 (61%)
Frame = +1
Query: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS 50
AIGID+GT+ V VW +VE++ N + + SYV F D+
Sbjct: 34 AIGIDIGTSQCSVAVWNGSQVELLKNTRNQKIMKSYVTFKDN 159
>TC233450
Length = 677
Score = 37.7 bits (86), Expect = 0.001
Identities = 16/65 (24%), Positives = 33/65 (50%)
Frame = +3
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNT 69
+G D+G + V + ++++ N + R TP+ V F++ +R+ G A M+ +T
Sbjct: 231 VGFDIGNENCVIAVVRQRGIDVLLNYESKRETPAVVCFSEKQRI*GSAGAASAMMHIKST 410
Query: 70 VFDAK 74
+ K
Sbjct: 411 ISQIK 425
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,473,638
Number of Sequences: 63676
Number of extensions: 20993
Number of successful extensions: 103
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 95
length of query: 74
length of database: 12,639,632
effective HSP length: 50
effective length of query: 24
effective length of database: 9,455,832
effective search space: 226939968
effective search space used: 226939968
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 52 (24.6 bits)
Medicago: description of AC144475.11


