

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.2 - phase: 0
(201 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
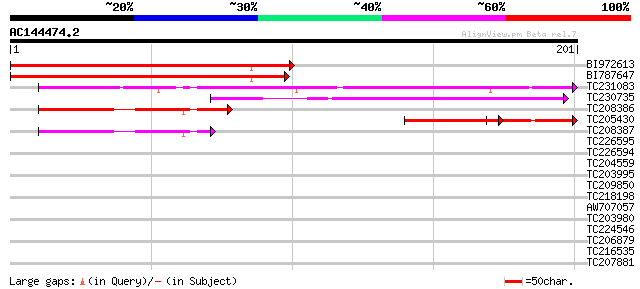
Score E
Sequences producing significant alignments: (bits) Value
BI972613 144 4e-35
BI787647 140 4e-34
TC231083 100 6e-22
TC230735 63 8e-11
TC208386 56 1e-08
TC205430 similar to UP|O22600 (O22600) Glycine-rich protein, par... 50 5e-07
TC208387 47 8e-06
TC226595 similar to UP|Q9SHG8 (Q9SHG8) SOUL-like protein, partia... 36 0.014
TC226594 weakly similar to UP|Q9SHG8 (Q9SHG8) SOUL-like protein,... 33 0.068
TC204559 similar to UP|Q76KW5 (Q76KW5) Class1 chitinase (Fragmen... 31 0.44
TC203995 similar to GB|AAP86659.1|32700018|AY230832 26S proteaso... 29 1.3
TC209850 weakly similar to UP|Q947C2 (Q947C2) P450 monooxygenase... 29 1.7
TC218198 similar to UP|Q9LXP3 (Q9LXP3) Protein kinase-like prote... 28 2.8
AW707057 28 2.8
TC203980 similar to GB|AAP86659.1|32700018|AY230832 26S proteaso... 28 3.7
TC224546 similar to UP|Q41707 (Q41707) Extensin class 1 protein ... 27 6.3
TC206879 similar to UP|Q8RY35 (Q8RY35) At2g44820/T13E15.17, part... 27 6.3
TC216535 27 8.3
TC207881 similar to UP|Q9FZM9 (Q9FZM9) Dihydroorotate dehydrogen... 27 8.3
>BI972613
Length = 423
Score = 144 bits (362), Expect = 4e-35
Identities = 70/105 (66%), Positives = 83/105 (78%), Gaps = 4/105 (3%)
Frame = +2
Query: 1 MGLVFGRFSAETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIG 60
MGLVFG+ + ET KYE++K+T Y IRKYAPS+VAE+TYDPS FKG+KDGGF +L +YIG
Sbjct: 104 MGLVFGKITVETAKYEVIKSTSEYEIRKYAPSVVAEVTYDPSQFKGNKDGGFMILANYIG 283
Query: 61 IFGKPQNTKTEKISMTTPVITKEN----KSSSEKIAMTVPVVTNE 101
GKPQNTK EKI+MT PVITK++ S EKIAMT PVVT E
Sbjct: 284 AVGKPQNTKPEKIAMTAPVITKDSVGGGGSDGEKIAMTAPVVTKE 418
>BI787647
Length = 423
Score = 140 bits (353), Expect = 4e-34
Identities = 68/100 (68%), Positives = 80/100 (80%), Gaps = 1/100 (1%)
Frame = +2
Query: 1 MGLVFGRFSAETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIG 60
MGLVFG+ S ET KYE++K+T Y IRKYAPS+VAE+TYDPS FKG+KDGGF +L +YIG
Sbjct: 119 MGLVFGKISVETAKYEVIKSTSEYEIRKYAPSVVAEVTYDPSQFKGNKDGGFMILANYIG 298
Query: 61 IFGKPQNTKTEKISMTTPVITKEN-KSSSEKIAMTVPVVT 99
GKPQNTK EKI+MT PVITK++ E IAMT PVVT
Sbjct: 299 AVGKPQNTKPEKIAMTAPVITKDSVGGDGETIAMTAPVVT 418
>TC231083
Length = 1086
Score = 100 bits (248), Expect = 6e-22
Identities = 75/199 (37%), Positives = 101/199 (50%), Gaps = 8/199 (4%)
Frame = +1
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGG---FKVLVDYIGIFGKPQN 67
ET +++L Y IR+ P VAE T P D +G F L +Y+ FGK N
Sbjct: 403 ETVDFKVLSRMDQYEIREVEPYFVAETTM-PGKSGFDFNGASRSFNALAEYL--FGK--N 567
Query: 68 TKTEKISMTTPVITKENKSSSEKIAMTVPVVTN--EKNKMVTMQFTLPSMYLKVEEVPKP 125
T EK+ MTTPV T +N+S K+ MT PV+T E M F +PS Y +P P
Sbjct: 568 TTKEKMEMTTPVFTSKNQSDGVKMDMTTPVLTTKMEDQDNWKMSFVMPSKY--GANLPLP 741
Query: 126 IDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDG---FKVIGDFLLGRYN 182
D V I+E K VV+F G +DE +K++ KLR L+ D K + +YN
Sbjct: 742 KDSSVRIKEVPRKIVAVVSFSGFVNDEEIKQRELKLRDALKSDSQFEIKEGTSVEVAQYN 921
Query: 183 PPAITIPMFRTNEVLIPVE 201
PP T+P R NE+ + VE
Sbjct: 922 PP-FTLPFQRRNEIALEVE 975
>TC230735
Length = 612
Score = 63.2 bits (152), Expect = 8e-11
Identities = 44/127 (34%), Positives = 61/127 (47%)
Frame = +3
Query: 72 KISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDERVV 131
+I MTTPV T+ N + K V++Q LP + + E +P P E V
Sbjct: 12 EIPMTTPVFTETNDADLSK---------------VSIQIVLP-LDKETESLPNPNQETVR 143
Query: 132 IREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAITIPMF 191
+R+ G V+ F G +++ V+EK + LR + KDG K LL RYN P T
Sbjct: 144 LRKVEGGIAAVMKFSGKPTEDTVREKEKTLRANIIKDGLKPQSGCLLARYNDPGRTWTFI 323
Query: 192 RTNEVLI 198
NEVLI
Sbjct: 324 MRNEVLI 344
>TC208386
Length = 757
Score = 56.2 bits (134), Expect = 1e-08
Identities = 35/70 (50%), Positives = 42/70 (60%), Gaps = 1/70 (1%)
Frame = +1
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIG-IFGKPQNTK 69
E+PKY+ILK T+NY +R+Y P +V E GDK G D G IFGK N+
Sbjct: 574 ESPKYQILKRTENYEVRQYNPFIVVET-------NGDKLSGSTGFNDVAGYIFGK--NST 726
Query: 70 TEKISMTTPV 79
TEKI MTTPV
Sbjct: 727 TEKIPMTTPV 756
>TC205430 similar to UP|O22600 (O22600) Glycine-rich protein, partial (19%)
Length = 421
Score = 50.4 bits (119), Expect = 5e-07
Identities = 24/35 (68%), Positives = 29/35 (82%)
Frame = +2
Query: 141 GVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGD 175
GV GVAS++VV+EKVEKLR LEKDGFKV+G+
Sbjct: 83 GVGNVSGVASEQVVREKVEKLRESLEKDGFKVVGE 187
Score = 43.1 bits (100), Expect = 9e-05
Identities = 22/32 (68%), Positives = 26/32 (80%)
Frame = +3
Query: 170 FKVIGDFLLGRYNPPAITIPMFRTNEVLIPVE 201
++ +G LLGRYNPP TIP FRTNEV+IPVE
Sbjct: 156 WRKMGLRLLGRYNPPW-TIPAFRTNEVMIPVE 248
>TC208387
Length = 487
Score = 46.6 bits (109), Expect = 8e-06
Identities = 30/64 (46%), Positives = 37/64 (56%), Gaps = 1/64 (1%)
Frame = +2
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIG-IFGKPQNTK 69
E+PKY+ILK T+NY +R+Y P +V E GDK G D G IFGK N+
Sbjct: 320 ESPKYQILKRTENYEVRQYNPFIVVET-------NGDKLSGSTGFNDVAGYIFGK--NST 472
Query: 70 TEKI 73
TEKI
Sbjct: 473 TEKI 484
>TC226595 similar to UP|Q9SHG8 (Q9SHG8) SOUL-like protein, partial (59%)
Length = 950
Score = 35.8 bits (81), Expect = 0.014
Identities = 22/73 (30%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Frame = +1
Query: 11 ETPKYEILKTTQNYVIRKY-APSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTK 69
E P Y+++ Y IR+Y +P ++ + GF+ L DYI + +N
Sbjct: 196 ECPSYDVIHVGNGYEIRRYNSPVWISNSPIQDISLVEATRTGFRRLFDYI----QGKNNY 363
Query: 70 TEKISMTTPVITK 82
+KI MT PVI++
Sbjct: 364 KQKIEMTAPVISE 402
>TC226594 weakly similar to UP|Q9SHG8 (Q9SHG8) SOUL-like protein, partial
(77%)
Length = 901
Score = 33.5 bits (75), Expect = 0.068
Identities = 22/73 (30%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Frame = +3
Query: 11 ETPKYEILKTTQNYVIRKY-APSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTK 69
E P Y+++ Y IR+Y +P ++ + GF+ L DYI + +N
Sbjct: 87 ECPSYDVIHFGNGYEIRRYNSPVWISNSPILDISLVEATRTGFRRLFDYI----QGKNNY 254
Query: 70 TEKISMTTPVITK 82
+KI MT PVI++
Sbjct: 255 KQKIEMTAPVISE 293
>TC204559 similar to UP|Q76KW5 (Q76KW5) Class1 chitinase (Fragment), partial
(97%)
Length = 1412
Score = 30.8 bits (68), Expect = 0.44
Identities = 24/69 (34%), Positives = 34/69 (48%)
Frame = -1
Query: 132 IREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAITIPMF 191
+++ GG GVV SD V +V LE G ++IG+ + N PA+T P F
Sbjct: 374 LKKRGGGDEGVVV---PKSDRVRHRRVPLGEQILELVGLEIIGNGEVTTVN*PALTFPSF 204
Query: 192 RTNEVLIPV 200
VL+PV
Sbjct: 203 VA--VLLPV 183
>TC203995 similar to GB|AAP86659.1|32700018|AY230832 26S proteasome subunit
RPN5a {Arabidopsis thaliana;} , partial (86%)
Length = 1250
Score = 29.3 bits (64), Expect = 1.3
Identities = 18/65 (27%), Positives = 35/65 (53%), Gaps = 9/65 (13%)
Frame = +3
Query: 115 MYLKVEEVPKPIDERVVIREEGG---------KKYGVVTFGGVASDEVVKEKVEKLRLCL 165
+Y+++E + I + I+EE G ++ V TFG +A E + +E++RLCL
Sbjct: 393 IYVEIERA-RLIKKLAKIKEEQGLIAEAADLMQEIAVETFGAMAKTEKIAFILEQVRLCL 569
Query: 166 EKDGF 170
++ +
Sbjct: 570 DRQDY 584
>TC209850 weakly similar to UP|Q947C2 (Q947C2) P450 monooxygenase, partial
(10%)
Length = 931
Score = 28.9 bits (63), Expect = 1.7
Identities = 16/48 (33%), Positives = 26/48 (53%)
Frame = +3
Query: 13 PKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIG 60
P Y+++ T+ +++ +PSLV EI D T ++D VLV G
Sbjct: 276 PIYKLMLGTKTFIVVS-SPSLVKEIVRDQDTVFANRDPPISVLVALYG 416
>TC218198 similar to UP|Q9LXP3 (Q9LXP3) Protein kinase-like protein, partial
(13%)
Length = 1230
Score = 28.1 bits (61), Expect = 2.8
Identities = 31/108 (28%), Positives = 45/108 (40%), Gaps = 11/108 (10%)
Frame = +3
Query: 63 GKPQNTKTEKISM---TTPVITKENKSSSEKIAMTVPVVTNEK-------NKMVTMQFTL 112
G + T E++S T PV+ SS + A TVPVVT K ++ VT+
Sbjct: 192 GDDKFTVRERLSPAAETVPVVATTKLSSQKPAAETVPVVTTNKLSSQKVLHEKVTLLQNP 371
Query: 113 PSMYLKVEEVPKPIDERV-VIREEGGKKYGVVTFGGVASDEVVKEKVE 159
+P D+ + VIR + V S++ VKE VE
Sbjct: 372 AQERHDTGHLPPAFDDVIHVIRHSSYR---------VGSEQPVKESVE 488
>AW707057
Length = 448
Score = 28.1 bits (61), Expect = 2.8
Identities = 20/47 (42%), Positives = 27/47 (56%)
Frame = +3
Query: 61 IFGKPQNTKTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVT 107
I KP++ +T+KIS T I+ KSSSE + P T EKN+ T
Sbjct: 297 ITSKPRS-RTKKISPQTSNISSSPKSSSETTTPS-PTTTCEKNQPFT 431
>TC203980 similar to GB|AAP86659.1|32700018|AY230832 26S proteasome subunit
RPN5a {Arabidopsis thaliana;} , partial (97%)
Length = 1723
Score = 27.7 bits (60), Expect = 3.7
Identities = 18/61 (29%), Positives = 33/61 (53%), Gaps = 9/61 (14%)
Frame = +2
Query: 115 MYLKVEEVPKPIDERVVIREEGG---------KKYGVVTFGGVASDEVVKEKVEKLRLCL 165
+Y+++E + I + I+EE G ++ V TFG +A E + +E++RLCL
Sbjct: 377 IYVEIERA-RLIKKLAKIKEEQGLIAEAADLMQEIAVETFGAMAKTEKIAFILEQVRLCL 553
Query: 166 E 166
+
Sbjct: 554 D 556
>TC224546 similar to UP|Q41707 (Q41707) Extensin class 1 protein precursor,
partial (67%)
Length = 1240
Score = 26.9 bits (58), Expect = 6.3
Identities = 29/124 (23%), Positives = 53/124 (42%), Gaps = 17/124 (13%)
Frame = -2
Query: 53 KVLVDYIGIFGKPQNTKTEKISMTTPVITKENKSSSEKIAMTVPV--------------- 97
KV+V + + G+ N+K E+ + V+ + EK+ + V
Sbjct: 480 KVVVGMVKVVGEIYNSKEEEGMVKVGVVIYNSMGEEEKVMVEVESDNSMGV*ELVKVVGE 301
Query: 98 VTNEKNKMVTMQFTLPS-MYLKVEEVPKPIDERVVIREEGGK-KYGVVTFGGVASDEVVK 155
+ N K + ++ + S + V E+ K + E +EE G K GVV + + +E V
Sbjct: 300 IYNSKEEEGMVKVGVESDNSMGV*ELVKVVGEIYNSKEEEGMVKVGVVIYNSMGEEEKVM 121
Query: 156 EKVE 159
+VE
Sbjct: 120 VEVE 109
>TC206879 similar to UP|Q8RY35 (Q8RY35) At2g44820/T13E15.17, partial (22%)
Length = 947
Score = 26.9 bits (58), Expect = 6.3
Identities = 18/71 (25%), Positives = 31/71 (43%)
Frame = +1
Query: 101 EKNKMVTMQFTLPSMYLKVEEVPKPIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEK 160
+KN+ + + P M + E K + ER+++ + GGK GG + K K E
Sbjct: 403 QKNQRLPLSVARPMMKKQKEREQKMLQERLILGQFGGK------LGGSSKRSAGKHKPEN 564
Query: 161 LRLCLEKDGFK 171
L + F+
Sbjct: 565 RGLKSSEGHFR 597
>TC216535
Length = 870
Score = 26.6 bits (57), Expect = 8.3
Identities = 17/58 (29%), Positives = 30/58 (51%)
Frame = +1
Query: 103 NKMVTMQFTLPSMYLKVEEVPKPIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEK 160
+K ++ Q + P YL VEE+P + RVV ++ G + + G V + ++ K K
Sbjct: 97 SKSLSQQVSYPP-YLFVEEIPNMGNSRVVTKDARGSQLTIFYGGQVLVFDDIQAKKAK 267
>TC207881 similar to UP|Q9FZM9 (Q9FZM9) Dihydroorotate dehydrogenase, partial
(53%)
Length = 1010
Score = 26.6 bits (57), Expect = 8.3
Identities = 15/36 (41%), Positives = 19/36 (52%)
Frame = +3
Query: 140 YGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGD 175
Y +GG A +K + L CLE+DGFK I D
Sbjct: 669 YTAFAYGGPALIPQIKAE---LAACLERDGFKSIVD 767
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.137 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,665,366
Number of Sequences: 63676
Number of extensions: 73164
Number of successful extensions: 389
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 379
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 381
length of query: 201
length of database: 12,639,632
effective HSP length: 92
effective length of query: 109
effective length of database: 6,781,440
effective search space: 739176960
effective search space used: 739176960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC144474.2


