

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.15 - phase: 0 /pseudo
(139 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
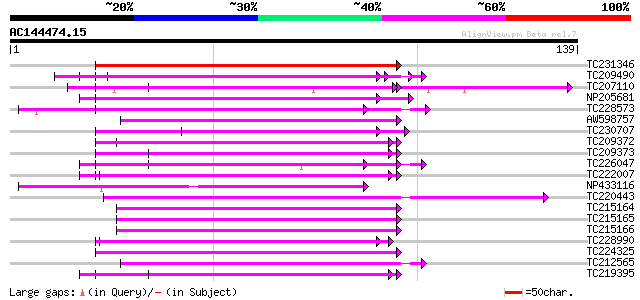
Score E
Sequences producing significant alignments: (bits) Value
TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2,... 60 4e-10
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 57 3e-09
TC207110 similar to UP|Q6V406 (Q6V406) Polygalacturonase inhibit... 51 2e-07
NP205681 receptor protein kinase-like protein 50 3e-07
TC228573 50 5e-07
AW598757 similar to GP|2982431|emb leucine rich repeat-like prot... 49 6e-07
TC230707 similar to UP|Q12937 (Q12937) BS-84, partial (4%) 47 4e-06
TC209372 weakly similar to UP|Q96477 (Q96477) LRR protein, parti... 46 5e-06
TC209373 weakly similar to UP|Q96477 (Q96477) LRR protein, parti... 46 5e-06
TC226047 weakly similar to UP|Q7XBG7 (Q7XBG7) Polygalacturonase-... 46 7e-06
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 45 1e-05
NP433116 candidate plant disease resistance protein [Glycine max] 45 1e-05
TC220443 weakly similar to UP|Q9S711 (Q9S711) ESTs C22657(S0014)... 44 2e-05
TC215164 homologue to UP|Q84N26 (Q84N26) Leucine-rich repeat pro... 44 3e-05
TC215165 homologue to UP|Q84N26 (Q84N26) Leucine-rich repeat pro... 44 3e-05
TC215166 homologue to UP|Q96477 (Q96477) LRR protein, partial (68%) 44 3e-05
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 44 3e-05
TC224325 weakly similar to UP|O49544 (O49544) Receptor protein k... 44 3e-05
TC212565 similar to UP|Q8LA44 (Q8LA44) Receptor protein kinase-l... 42 7e-05
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 42 7e-05
>TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2, partial
(14%)
Length = 1790
Score = 59.7 bits (143), Expect = 4e-10
Identities = 30/75 (40%), Positives = 47/75 (62%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L++++ L + + LSGPL SL +L++L V+ L N F+ P+P FAN +L TLNL
Sbjct: 52 LQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIPSPFANLSSLRTLNLAHN 231
Query: 82 GLIGTFPQNIFQIKS 96
L GT P++ +K+
Sbjct: 232 RLNGTIPKSFEFLKN 276
Score = 37.4 bits (85), Expect = 0.002
Identities = 24/84 (28%), Positives = 42/84 (49%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L+ L++S LSG + + + +++ L + L NN S +PQ+ ++ L+ LNL
Sbjct: 1486 LSALRFLNLSRNHLSGEIPNDMGKMKLLESLDLSLNNISGQIPQSLSDLSFLSFLNLSYH 1665
Query: 82 GLIGTFPQNIFQIKSQLLTYLTTP 105
L G P + + L+Y P
Sbjct: 1666 NLSGRIPTSTQLQSFEELSYTGNP 1737
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 57.0 bits (136), Expect = 3e-09
Identities = 31/91 (34%), Positives = 52/91 (57%)
Frame = +1
Query: 12 GHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFK 71
GH +LP+ L+ L + D + +GPL L +LE L + L N FS +P++++ FK
Sbjct: 493 GHFPGQIILPMTKLEVLDVYDNNFTGPLPVELVKLEKLKYLKLDGNYFSGSIPESYSEFK 672
Query: 72 NLTTLNLRKCGLIGTFPQNIFQIKSQLLTYL 102
+L L+L L G P+++ ++K+ L YL
Sbjct: 673 SLEFLSLSTNSLSGKIPKSLSKLKT--LRYL 759
Score = 46.6 bits (109), Expect = 4e-06
Identities = 24/78 (30%), Positives = 41/78 (51%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
++ L+ L +S C+LSG + SL L NL + L NN + +P + +L +L+L
Sbjct: 814 MKSLRYLDLSSCNLSGEIPPSLANLTNLDTLFLQINNLTGTIPSELSAMVSLMSLDLSIN 993
Query: 82 GLIGTFPQNIFQIKSQLL 99
L G P + Q+++ L
Sbjct: 994 DLTGEIPMSFSQLRNLTL 1047
Score = 43.1 bits (100), Expect = 4e-05
Identities = 25/74 (33%), Positives = 38/74 (50%)
Frame = +1
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
AL LR LQ LS+ + G + + L LTV+ + NN + P+P T +LT ++
Sbjct: 1519 ALKNLRALQTLSLDANEFVGEIPGEVFDLPMLTVVNISGNNLTGPIPTTLTRCVSLTAVD 1698
Query: 78 LRKCGLIGTFPQNI 91
L + L G P+ I
Sbjct: 1699 LSRNMLEGKIPKGI 1740
Score = 39.3 bits (90), Expect = 6e-04
Identities = 23/69 (33%), Positives = 32/69 (46%)
Frame = +1
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L +S DL+G + S ++L NLT++ +NN VP NL TL L
Sbjct: 967 LMSLDLSINDLTGEIPMSFSQLRNLTLMNFFQNNLRGSVPSFVGELPNLETLQLWDNNFS 1146
Query: 85 GTFPQNIFQ 93
P N+ Q
Sbjct: 1147 FVLPPNLGQ 1173
Score = 38.1 bits (87), Expect = 0.001
Identities = 19/67 (28%), Positives = 36/67 (53%)
Frame = +1
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L +++S +L+GP+ ++LTR +LT + L N +P+ N +L+ N+ +
Sbjct: 1612 LTVVNISGNNLTGPIPTTLTRCVSLTAVDLSRNMLEGKIPKGIKNLTDLSIFNVSINQIS 1791
Query: 85 GTFPQNI 91
G P+ I
Sbjct: 1792 GPVPEEI 1812
Score = 34.3 bits (77), Expect = 0.020
Identities = 19/64 (29%), Positives = 30/64 (46%)
Frame = +1
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L + +S L G + + L +L++ + N S PVP+ +LTTL+L I
Sbjct: 1684 LTAVDLSRNMLEGKIPKGIKNLTDLSIFNVSINQISGPVPEEIRFMLSLTTLDLSNNNFI 1863
Query: 85 GTFP 88
G P
Sbjct: 1864 GKVP 1875
Score = 31.2 bits (69), Expect = 0.17
Identities = 23/87 (26%), Positives = 39/87 (44%), Gaps = 1/87 (1%)
Frame = +1
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTR-LENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
L L L+ L++S SG + + L V+ + +NNF+ P+P + L L
Sbjct: 439 LAALTSLKHLNISHNVFSGHFPGQIILPMTKLEVLDVYDNNFTGPLPVELVKLEKLKYLK 618
Query: 78 LRKCGLIGTFPQNIFQIKSQLLTYLTT 104
L G+ P++ + KS L+T
Sbjct: 619 LDGNYFSGSIPESYSEFKSLEFLSLST 699
Score = 30.0 bits (66), Expect = 0.37
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 10/64 (15%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVP----------QTFANFK 71
L DL ++S +SGP+ + + +LT + L NNF VP ++FA
Sbjct: 1747 LTDLSIFNVSINQISGPVPEEIRFMLSLTTLDLSNNNFIGKVPTGGQFAVFSEKSFAGNP 1926
Query: 72 NLTT 75
NL T
Sbjct: 1927 NLCT 1938
>TC207110 similar to UP|Q6V406 (Q6V406) Polygalacturonase inhibiting protein,
partial (93%)
Length = 1165
Score = 51.2 bits (121), Expect = 2e-07
Identities = 42/138 (30%), Positives = 69/138 (49%), Gaps = 14/138 (10%)
Frame = +3
Query: 15 WSNALLPLRD-------LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTF 67
W+N P+ D LQ L +S +LSGP+ SSL++L NL + L N + P+P++F
Sbjct: 387 WTNVSGPVPDFLARLTNLQFLDLSFNNLSGPIPSSLSQLSNLVSLRLDRNRLTGPIPESF 566
Query: 68 ANFKNL-TTLNLRKCGLIGTFPQNIFQIKSQLLTY----LTTPTSMVF--FQTTHLVNLF 120
+FK +L L L G P ++ I Q + + L S++F +TT +V++
Sbjct: 567 GSFKKPGPSLWLSHNQLSGPIPASLANIDPQRIDFSRNKLEGDASVLFGRNKTTQIVDVS 746
Query: 121 IALYCVTLSSLEHVHTLL 138
LS +E +L+
Sbjct: 747 RNSLAFDLSKVEFPTSLI 800
Score = 48.5 bits (114), Expect = 1e-06
Identities = 25/74 (33%), Positives = 40/74 (53%)
Frame = +3
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L+++ +S ++SGP+ L RL NL + L NN S P+P + + NL +L L +
Sbjct: 357 LTKLKEIYISWTNVSGPVPDFLARLTNLQFLDLSFNNLSGPIPSSLSQLSNLVSLRLDRN 536
Query: 82 GLIGTFPQNIFQIK 95
L G P++ K
Sbjct: 537 RLTGPIPESFGSFK 578
Score = 39.3 bits (90), Expect = 6e-04
Identities = 20/62 (32%), Positives = 31/62 (49%)
Frame = +3
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
L+GP+ ++ +L L I + N S PVP A NL L+L L G P ++ Q+
Sbjct: 324 LTGPIQPTIAKLTKLKEIYISWTNVSGPVPDFLARLTNLQFLDLSFNNLSGPIPSSLSQL 503
Query: 95 KS 96
+
Sbjct: 504 SN 509
>NP205681 receptor protein kinase-like protein
Length = 2946
Score = 50.1 bits (118), Expect = 3e-07
Identities = 25/78 (32%), Positives = 41/78 (52%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
+ L+ L +S C+LSG + SL + NL + L NN + +P ++ +L +L+L
Sbjct: 724 MESLKYLDLSSCNLSGEIPPSLANMRNLDTLFLQMNNLTGTIPSELSDMVSLMSLDLSFN 903
Query: 82 GLIGTFPQNIFQIKSQLL 99
GL G P Q+K+ L
Sbjct: 904 GLTGEIPTRFSQLKNLTL 957
Score = 42.7 bits (99), Expect = 6e-05
Identities = 24/74 (32%), Positives = 38/74 (50%)
Frame = +1
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
AL LR LQ LS+ + G + + L LTV+ + NN + P+P TF +L ++
Sbjct: 1429 ALKNLRALQTLSLDTNEFLGEIPGEVFDLPMLTVVNISGNNLTGPIPTTFTRCVSLAAVD 1608
Query: 78 LRKCGLIGTFPQNI 91
L + L G P+ +
Sbjct: 1609 LSRNMLDGEIPKGM 1650
Score = 37.4 bits (85), Expect = 0.002
Identities = 24/83 (28%), Positives = 41/83 (48%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L++L ++ +L G + S + L NL + L ENNFSS +PQ ++ K
Sbjct: 940 LKNLTLMNFFHNNLRGSVPSFVGELPNLETLQLWENNFSSELPQNLGQNGKFKFFDVTKN 1119
Query: 82 GLIGTFPQNIFQIKSQLLTYLTT 104
G P+++ + +L T+L T
Sbjct: 1120 HFSGLIPRDLCK-SGRLQTFLIT 1185
>TC228573
Length = 1628
Score = 49.7 bits (117), Expect = 5e-07
Identities = 35/104 (33%), Positives = 53/104 (50%), Gaps = 3/104 (2%)
Frame = +3
Query: 3 LDG---ISITSRGHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNF 59
LDG I++ + + N L L +LQ + + + +LSG + ++ L L V+ L N F
Sbjct: 351 LDGLYLINLINISGPFPNFLFQLPNLQFIYLENNNLSGRIPDNIGNLTRLDVLSLTGNRF 530
Query: 60 SSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIKSQLLTYLT 103
PVP + LT L L L GT PQ I ++ + LTYL+
Sbjct: 531 IGPVPSSITKLTQLTQLKLGNNFLTGTVPQGIAKLVN--LTYLS 656
Score = 46.6 bits (109), Expect = 4e-06
Identities = 28/67 (41%), Positives = 36/67 (52%)
Frame = +3
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L LS++ GP+ SS+T+L LT + LG N + VPQ A NLT L+L
Sbjct: 489 LTRLDVLSLTGNRFIGPVPSSITKLTQLTQLKLGNNFLTGTVPQGIAKLVNLTYLSLEGN 668
Query: 82 GLIGTFP 88
L GT P
Sbjct: 669 QLEGTIP 689
Score = 37.0 bits (84), Expect = 0.003
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Frame = +3
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L + LSG + L + + L + L N FS VP +F N + LNL L+
Sbjct: 789 LTYLELGHNSLSGKIPDFLGKFKALDTLDLSWNKFSGTVPASFKNLTKIFNLNLSNNLLV 968
Query: 85 GTFPQ-NIFQIKS 96
FP+ N+ I+S
Sbjct: 969 DPFPEMNVKGIES 1007
>AW598757 similar to GP|2982431|emb leucine rich repeat-like protein
{Arabidopsis thaliana}, partial (6%)
Length = 415
Score = 49.3 bits (116), Expect = 6e-07
Identities = 25/69 (36%), Positives = 38/69 (54%)
Frame = +2
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
L++S+ LSG + +SL RL+NL + L N S P+P T +N +L +L L L G
Sbjct: 179 LNLSESSLSGSISTSLGRLQNLIHLDLSSNRLSGPIPPTLSNLTSLESLLLHSNQLTGQI 358
Query: 88 PQNIFQIKS 96
P + + S
Sbjct: 359 PTELHSLTS 385
>TC230707 similar to UP|Q12937 (Q12937) BS-84, partial (4%)
Length = 990
Score = 46.6 bits (109), Expect = 4e-06
Identities = 23/70 (32%), Positives = 38/70 (53%)
Frame = +3
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L L ++D + GP+ SS++ L NL + L N+FS +P + K+L +L+L
Sbjct: 12 LTQLTTLDLADNNFFGPIPSSISLLSNLQTLTLRSNSFSGTIPPSITTLKSLLSLDLAHN 191
Query: 82 GLIGTFPQNI 91
L G P ++
Sbjct: 192SLSGYLPNSM 221
Score = 45.8 bits (107), Expect = 7e-06
Identities = 22/56 (39%), Positives = 32/56 (56%)
Frame = +3
Query: 43 LTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIKSQL 98
+++L LT + L +NNF P+P + + NL TL LR GT P +I +KS L
Sbjct: 3 ISQLTQLTTLDLADNNFFGPIPSSISLLSNLQTLTLRSNSFSGTIPPSITTLKSLL 170
>TC209372 weakly similar to UP|Q96477 (Q96477) LRR protein, partial (71%)
Length = 690
Score = 46.2 bits (108), Expect = 5e-06
Identities = 24/73 (32%), Positives = 38/73 (51%)
Frame = +2
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ LQ L + DL GP+ L L+NL + L +NN + +P T +N N+ L L
Sbjct: 293 LQRLQFLELYKNDLMGPIPKELGELKNLLSLGLYQNNLTGSIPATLSNLSNIKFLRLNSN 472
Query: 82 GLIGTFPQNIFQI 94
L G P+ + ++
Sbjct: 473 KLTGRIPRELTKL 511
Score = 41.6 bits (96), Expect = 1e-04
Identities = 21/70 (30%), Positives = 35/70 (50%)
Frame = +2
Query: 27 KLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGT 86
+L + LSG L L RL+ L + L +N+ P+P+ KNL +L L + L G+
Sbjct: 236 RLDLGHAKLSGHLVPELGRLQRLQFLELYKNDLMGPIPKELGELKNLLSLGLYQNNLTGS 415
Query: 87 FPQNIFQIKS 96
P + + +
Sbjct: 416 IPATLSNLSN 445
>TC209373 weakly similar to UP|Q96477 (Q96477) LRR protein, partial (51%)
Length = 654
Score = 46.2 bits (108), Expect = 5e-06
Identities = 24/73 (32%), Positives = 38/73 (51%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ LQ L + DL GP+ L L+NL + L +NN + +P T +N N+ L L
Sbjct: 106 LQRLQFLELYKNDLMGPIPKELGELKNLLSLGLYQNNLTGSIPATLSNLSNIKFLRLNSN 285
Query: 82 GLIGTFPQNIFQI 94
L G P+ + ++
Sbjct: 286 KLTGRIPRELTKL 324
Score = 39.3 bits (90), Expect = 6e-04
Identities = 20/62 (32%), Positives = 32/62 (51%)
Frame = +1
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
LSG L L RL+ L + L +N+ P+P+ KNL +L L + L G+ P + +
Sbjct: 73 LSGHLVPELGRLQRLQFLELYKNDLMGPIPKELGELKNLLSLGLYQNNLTGSIPATLSNL 252
Query: 95 KS 96
+
Sbjct: 253 SN 258
>TC226047 weakly similar to UP|Q7XBG7 (Q7XBG7) Polygalacturonase-inhibiting
protein, partial (58%)
Length = 1342
Score = 45.8 bits (107), Expect = 7e-06
Identities = 24/71 (33%), Positives = 38/71 (52%)
Frame = +1
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
A+ L L+ L +S+ +LSGP+ +L+NL I + NN S P+P + L L+
Sbjct: 397 AIAKLTKLKYLDLSNNNLSGPIPDFFAQLKNLDDIDISFNNLSGPIPSSLGKLPKLAYLD 576
Query: 78 LRKCGLIGTFP 88
L + L G+ P
Sbjct: 577 LSRNKLTGSIP 609
Score = 42.7 bits (99), Expect = 6e-05
Identities = 22/68 (32%), Positives = 35/68 (51%)
Frame = +1
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
L GP+ ++ +L L + L NN S P+P FA KNL +++ L G P ++ ++
Sbjct: 376 LVGPIQPAIAKLTKLKYLDLSNNNLSGPIPDFFAQLKNLDDIDISFNNLSGPIPSSLGKL 555
Query: 95 KSQLLTYL 102
L YL
Sbjct: 556 PK--LAYL 573
Score = 40.0 bits (92), Expect = 4e-04
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANF-KNLTTLNLRK 80
L++L + +S +LSGP+ SSL +L L + L N + +P +F +F K + L K
Sbjct: 481 LKNLDDIDISFNNLSGPIPSSLGKLPKLAYLDLSRNKLTGSIPASFGSFQKPGPAIMLSK 660
Query: 81 CGLIGTFPQNIFQIKS 96
L G P ++ + S
Sbjct: 661 NQLSGRLPASLANLDS 708
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 45.1 bits (105), Expect = 1e-05
Identities = 23/75 (30%), Positives = 38/75 (50%)
Frame = +3
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L K+ S SGP+ +++ + LT + L N S +P + L LNL K
Sbjct: 1512 LQQLSKIDFSGNKFSGPIAPEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLNLSKN 1691
Query: 82 GLIGTFPQNIFQIKS 96
L+G+ P +I ++S
Sbjct: 1692 HLVGSIPSSISSMQS 1736
Score = 44.3 bits (103), Expect = 2e-05
Identities = 23/74 (31%), Positives = 39/74 (52%)
Frame = +3
Query: 23 RDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCG 82
R + L+++ DLSG L + + L L+ + L N FS P+P + + L LNL
Sbjct: 216 RHVTALNLTGLDLSGTLSADVAHLPFLSNLSLAANKFSGPIPPSLSALSGLRYLNLSNNV 395
Query: 83 LIGTFPQNIFQIKS 96
TFP +++++S
Sbjct: 396 FNETFPSELWRLQS 437
Score = 42.4 bits (98), Expect = 7e-05
Identities = 27/77 (35%), Positives = 39/77 (50%)
Frame = +3
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
AL L+ L L + LSG L L L++L + L N S +P +F KN+T LN
Sbjct: 780 ALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPASFGELKNITLLN 959
Query: 78 LRKCGLIGTFPQNIFQI 94
L + L G P+ I ++
Sbjct: 960 LFRNKLHGAIPEFIGEL 1010
Score = 37.4 bits (85), Expect = 0.002
Identities = 18/70 (25%), Positives = 36/70 (50%)
Frame = +3
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L+ + +S+ LSG + +S L+N+T++ L N +P+ L + L +
Sbjct: 864 LKSLKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWEN 1043
Query: 82 GLIGTFPQNI 91
L G+ P+ +
Sbjct: 1044NLTGSIPEGL 1073
Score = 37.0 bits (84), Expect = 0.003
Identities = 20/55 (36%), Positives = 29/55 (52%)
Frame = +3
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQ 89
L GP+ SL E+LT I +GEN + +P+ LT + L+ L G FP+
Sbjct: 1191 LFGPIPESLGTCESLTRIRMGENFLNGSIPKGLFGLPKLTQVELQDNYLSGEFPE 1355
>NP433116 candidate plant disease resistance protein [Glycine max]
Length = 686
Score = 44.7 bits (104), Expect = 1e-05
Identities = 32/91 (35%), Positives = 46/91 (50%), Gaps = 5/91 (5%)
Frame = +1
Query: 3 LDGISITSRGHEWSNALLP-----LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGEN 57
LD + I S + LP L LQ L + +LSG + +SL+ L V+ L N
Sbjct: 370 LDAVKIISLRSNLLSGNLPADIGSLPSLQYLYLQHNNLSGDIPASLS--PQLIVLDLSYN 543
Query: 58 NFSSPVPQTFANFKNLTTLNLRKCGLIGTFP 88
+F+ +P+TF N LT+LNL+ L G P
Sbjct: 544 SFTGVIPKTFQNMSVLTSLNLQNNSLSGQIP 636
>TC220443 weakly similar to UP|Q9S711 (Q9S711) ESTs C22657(S0014)
(Transmembrane protein kinase), partial (8%)
Length = 634
Score = 44.3 bits (103), Expect = 2e-05
Identities = 31/109 (28%), Positives = 49/109 (44%)
Frame = +3
Query: 24 DLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
++ + +S SG + S ++ NL V +NNF+ +P+ LTTL L + L
Sbjct: 150 NISRFEISYNQFSGGIPSGVSSWTNLVVFDASKNNFNGSIPRQLTALPKLTTLLLDQNQL 329
Query: 84 IGTFPQNIFQIKSQLLTYLTTPTSMVFFQTTHLVNLFIALYCVTLSSLE 132
G P +I KS L L + ++ Q H + AL + LS E
Sbjct: 330 TGELPSDIISWKS--LVALNLSQNQLYGQIPHAIGQLPALSQLDLSENE 470
>TC215164 homologue to UP|Q84N26 (Q84N26) Leucine-rich repeat protein,
partial (55%)
Length = 868
Score = 43.9 bits (102), Expect = 3e-05
Identities = 21/70 (30%), Positives = 40/70 (57%)
Frame = +2
Query: 27 KLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGT 86
++ + + +LSG L L +LE+L + L +NN +P N K+L +L+L + GT
Sbjct: 383 RVDLGNSNLSGHLVPELGKLEHLQYLELYKNNIQGTIPPELGNLKSLVSLDLYNNNISGT 562
Query: 87 FPQNIFQIKS 96
P ++ ++K+
Sbjct: 563 IPPSLGKLKN 592
>TC215165 homologue to UP|Q84N26 (Q84N26) Leucine-rich repeat protein,
partial (84%)
Length = 1129
Score = 43.9 bits (102), Expect = 3e-05
Identities = 21/70 (30%), Positives = 40/70 (57%)
Frame = +3
Query: 27 KLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGT 86
++ + + +LSG L L +LE+L + L +NN +P N K+L +L+L + GT
Sbjct: 447 RVDLGNSNLSGHLVPELGKLEHLQYLELYKNNIQGTIPPELGNLKSLVSLDLYNNNISGT 626
Query: 87 FPQNIFQIKS 96
P ++ ++K+
Sbjct: 627 IPPSLGKLKN 656
>TC215166 homologue to UP|Q96477 (Q96477) LRR protein, partial (68%)
Length = 689
Score = 43.9 bits (102), Expect = 3e-05
Identities = 21/70 (30%), Positives = 40/70 (57%)
Frame = +1
Query: 27 KLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGT 86
++ + + +LSG L L +LE+L + L +NN +P N K+L +L+L + GT
Sbjct: 16 RVDLGNSNLSGHLVPELGKLEHLQYLELYKNNIQGTIPPELGNLKSLVSLDLYNNNISGT 195
Query: 87 FPQNIFQIKS 96
P ++ ++K+
Sbjct: 196IPPSLGKLKN 225
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 43.9 bits (102), Expect = 3e-05
Identities = 27/73 (36%), Positives = 39/73 (52%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L++L L + LSG L L L++L + L N S VP +FA KNLT LNL +
Sbjct: 859 LQNLDTLFLQVNALSGSLTPELGSLKSLKSMDLSNNMLSGEVPASFAELKNLTLLNLFRN 1038
Query: 82 GLIGTFPQNIFQI 94
L G P+ + ++
Sbjct: 1039KLHGAIPEFVGEL 1077
Score = 43.9 bits (102), Expect = 3e-05
Identities = 23/69 (33%), Positives = 35/69 (50%)
Frame = +1
Query: 23 RDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCG 82
R + L+++ LSG L L+ L L+ + L +N FS P+P +F+ L LNL
Sbjct: 283 RHVTSLNLTSLSLSGTLSDDLSHLPFLSHLSLADNKFSGPIPASFSALSALRFLNLSNNV 462
Query: 83 LIGTFPQNI 91
TFP +
Sbjct: 463 FNATFPSQL 489
Score = 41.2 bits (95), Expect = 2e-04
Identities = 23/70 (32%), Positives = 36/70 (50%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L++L L++ L G + + L L V+ L ENNF+ +PQ N LT ++L
Sbjct: 1003 LKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFTGSIPQNLGNNGRLTLVDLSSN 1182
Query: 82 GLIGTFPQNI 91
+ GT P N+
Sbjct: 1183 KITGTLPPNM 1212
Score = 41.2 bits (95), Expect = 2e-04
Identities = 21/70 (30%), Positives = 36/70 (51%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L+ + +S+ LSG + +S L+NLT++ L N +P+ L L L +
Sbjct: 931 LKSLKSMDLSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWEN 1110
Query: 82 GLIGTFPQNI 91
G+ PQN+
Sbjct: 1111 NFTGSIPQNL 1140
Score = 35.0 bits (79), Expect = 0.012
Identities = 18/56 (32%), Positives = 30/56 (53%)
Frame = +1
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQN 90
L GP+ SL + ++L I +GEN + +P+ LT + L+ L G FP++
Sbjct: 1258 LFGPIPDSLGKCKSLNRIRMGENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPED 1425
>TC224325 weakly similar to UP|O49544 (O49544) Receptor protein kinase -
like protein, partial (7%)
Length = 424
Score = 43.5 bits (101), Expect = 3e-05
Identities = 22/75 (29%), Positives = 40/75 (53%)
Frame = +3
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ LQ ++ +L+G + S+ L +L + +G NN +PQ + KNL+ +++
Sbjct: 15 LQKLQYFYVAKNNLTGEVPPSIGNLSSLIELSVGLNNLEGKIPQEVCSLKNLSLMSVPVN 194
Query: 82 GLIGTFPQNIFQIKS 96
L GT P +F + S
Sbjct: 195KLSGTIPSEVFSLSS 239
Score = 26.9 bits (58), Expect = 3.2
Identities = 13/43 (30%), Positives = 25/43 (57%)
Frame = +3
Query: 17 NALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNF 59
N + L++L+K+ +S+ LSG + S+ +L + L N+F
Sbjct: 291 NVVSKLKNLEKMDVSENHLSGDIPGSIGDCTSLEYLYLQGNSF 419
>TC212565 similar to UP|Q8LA44 (Q8LA44) Receptor protein kinase-like protein,
partial (24%)
Length = 1021
Score = 42.4 bits (98), Expect = 7e-05
Identities = 25/75 (33%), Positives = 36/75 (47%)
Frame = +2
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
L + +LSG L S+ L NL +VL NN + P+P L TL+L G
Sbjct: 728 LGIPSQNLSGTLSPSIGNLTNLQTVVLQNNNITGPIPSEIGKLSKLQTLDLSDNFFSGEI 907
Query: 88 PQNIFQIKSQLLTYL 102
P ++ ++S L YL
Sbjct: 908 PPSMGHLRS--LQYL 946
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 42.4 bits (98), Expect = 7e-05
Identities = 22/75 (29%), Positives = 37/75 (49%)
Frame = +3
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L K+ S SGP+ +++ + LT + L N S +P + L LNL +
Sbjct: 1263 LQQLSKIDFSGNKFSGPIVPEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLNLSRN 1442
Query: 82 GLIGTFPQNIFQIKS 96
L+G P +I ++S
Sbjct: 1443 HLVGGIPSSISSMQS 1487
Score = 41.6 bits (96), Expect = 1e-04
Identities = 27/77 (35%), Positives = 38/77 (49%)
Frame = +3
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
AL L+ L L + LSG L L L++L + L N S +P F KN+T LN
Sbjct: 531 ALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPARFGELKNITLLN 710
Query: 78 LRKCGLIGTFPQNIFQI 94
L + L G P+ I ++
Sbjct: 711 LFRNKLHGAIPEFIGEL 761
Score = 39.7 bits (91), Expect = 5e-04
Identities = 20/62 (32%), Positives = 32/62 (51%)
Frame = +3
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
LSGPL + + L L+ + L N FS P+P + + L LNL TFP + ++
Sbjct: 3 LSGPLSADVAHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRL 182
Query: 95 KS 96
++
Sbjct: 183QN 188
Score = 37.0 bits (84), Expect = 0.003
Identities = 20/55 (36%), Positives = 29/55 (52%)
Frame = +3
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQ 89
L GP+ SL E+LT I +GEN + +P+ LT + L+ L G FP+
Sbjct: 942 LFGPIPESLGSCESLTRIRMGENFLNGSIPRGLFGLPKLTQVELQDNYLSGEFPE 1106
Score = 33.9 bits (76), Expect = 0.026
Identities = 16/70 (22%), Positives = 34/70 (47%)
Frame = +3
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L+ + +S+ LSG + + L+N+T++ L N +P+ L + L +
Sbjct: 615 LKSLKSMDLSNNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWEN 794
Query: 82 GLIGTFPQNI 91
G+ P+ +
Sbjct: 795 NFTGSIPEGL 824
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,090,868
Number of Sequences: 63676
Number of extensions: 74125
Number of successful extensions: 623
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 552
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 618
length of query: 139
length of database: 12,639,632
effective HSP length: 87
effective length of query: 52
effective length of database: 7,099,820
effective search space: 369190640
effective search space used: 369190640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC144474.15


