

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144459.2 - phase: 0 /pseudo
(824 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
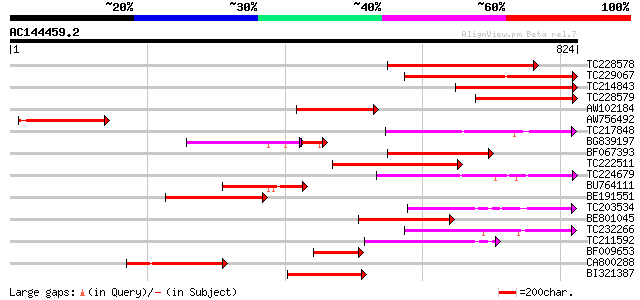
Score E
Sequences producing significant alignments: (bits) Value
TC228578 similar to GB|CAC05466.1|9955527|ATT5E8 potassium trans... 388 e-108
TC229067 288 7e-78
TC214843 similar to GB|CAC05466.1|9955527|ATT5E8 potassium trans... 286 3e-77
TC228579 similar to UP|PT13_ARATH (Q8LPL8) Potassium transporter... 244 1e-64
AW102184 similar to GP|9955527|emb potassium transport protein-l... 223 3e-58
AW756492 similar to GP|9955527|emb| potassium transport protein-... 221 1e-57
TC217848 similar to UP|PT11_ARATH (O64769) Potassium transporter... 200 2e-51
BG839197 160 2e-46
BF067393 183 2e-46
TC222511 similar to UP|Q94KC5 (Q94KC5) Potassium transporter HAK... 171 2e-42
TC224679 similar to UP|Q94KC6 (Q94KC6) Potassium transporter HAK... 169 4e-42
BU764111 similar to GP|9955527|emb potassium transport protein-l... 163 3e-40
BE191551 152 8e-37
TC203534 151 1e-36
BE801045 147 1e-35
TC232266 similar to UP|Q94KC5 (Q94KC5) Potassium transporter HAK... 145 5e-35
TC211592 similar to UP|POT6_ARATH (Q8W4I4) Potassium transporter... 144 1e-34
BF009653 similar to GP|9955527|emb| potassium transport protein-... 130 2e-30
CA800288 126 4e-29
BI321387 123 3e-28
>TC228578 similar to GB|CAC05466.1|9955527|ATT5E8 potassium transport
protein-like {Arabidopsis thaliana;} , partial (22%)
Length = 670
Score = 388 bits (997), Expect = e-108
Identities = 196/222 (88%), Positives = 207/222 (92%), Gaps = 2/222 (0%)
Frame = +1
Query: 549 GSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIH 608
GSKLKYETEVKQKLS DLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPA+H
Sbjct: 4 GSKLKYETEVKQKLSMDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAVH 183
Query: 609 SMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLME 668
SMIIFVSIKYVPV MVPQSERFLFRRVCQRSYH+FRCIARYGYKD RKENHQ FEQLLME
Sbjct: 184 SMIIFVSIKYVPVPMVPQSERFLFRRVCQRSYHIFRCIARYGYKDVRKENHQTFEQLLME 363
Query: 669 SLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLADFNESF--M 726
SLEKFIRREAQERSLES+GD+DT+ EDEY+GSRVLIAPNGSVYSLGVPLLADFN++ +
Sbjct: 364 SLEKFIRREAQERSLESEGDDDTDSEDEYSGSRVLIAPNGSVYSLGVPLLADFNDTTIPI 543
Query: 727 PSFEPSTSEEAGPPSPKPLVLDAEQLLERELSFIRNAKESGL 768
P+FE STSEEA P SPKP VLDAEQ LERELS IR AKESG+
Sbjct: 544 PNFEASTSEEANPESPKPPVLDAEQSLERELSCIRKAKESGV 669
>TC229067
Length = 1161
Score = 288 bits (737), Expect = 7e-78
Identities = 146/252 (57%), Positives = 183/252 (71%), Gaps = 2/252 (0%)
Frame = +3
Query: 575 GTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRR 634
GT+R PGIGLLYNELV+GIP IF FL LPA+HS I+FV IKYVPV +VPQ ERFLFRR
Sbjct: 6 GTVRVPGIGLLYNELVQGIPSIFLQFLLNLPALHSTIVFVCIKYVPVPVVPQEERFLFRR 185
Query: 635 VCQRSYHLFRCIARYGYKDARKENHQAFEQLLMESLEKFIRREAQERSLESDGDEDTELE 694
VC + YH+FRC+ARYGYKD RKE+H AFEQLL+ESLEKF+RREA E +LE +G+ E++
Sbjct: 186 VCPKDYHIFRCVARYGYKDVRKEDHHAFEQLLIESLEKFLRREALETALELEGNLSDEMD 365
Query: 695 DEYAGSRVLIAP-NGSVYSLGVPLLADFNESFMPSFEPSTSEEAGPPSPKP-LVLDAEQL 752
+RV P + + L +PL+ D + + S S+E P + D +
Sbjct: 366 SVSVNTRVSDVPVDTTAEELRIPLVHD--QKLEEAGASSASQEVASALPSSYMSSDEDPA 539
Query: 753 LERELSFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVP 812
LE ELS +R A ESG YLLGHGD+RA+K+S+F KKL+INYFYAFLRKNCR G N+ VP
Sbjct: 540 LEYELSALREALESGFTYLLGHGDVRAKKNSFFFKKLMINYFYAFLRKNCRGGTANMRVP 719
Query: 813 HSHLMQVGMTYM 824
H++++QVGMTYM
Sbjct: 720 HTNIIQVGMTYM 755
>TC214843 similar to GB|CAC05466.1|9955527|ATT5E8 potassium transport
protein-like {Arabidopsis thaliana;} , partial (18%)
Length = 1749
Score = 286 bits (731), Expect = 3e-77
Identities = 143/177 (80%), Positives = 151/177 (84%)
Frame = +3
Query: 648 RYGYKDARKENHQAFEQLLMESLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPN 707
RYGYKD RKENHQ FEQLL+ESLEKFIRREAQERSLESDGD+DT+ EDEY SRVLIAPN
Sbjct: 90 RYGYKDVRKENHQTFEQLLIESLEKFIRREAQERSLESDGDDDTDSEDEYPNSRVLIAPN 269
Query: 708 GSVYSLGVPLLADFNESFMPSFEPSTSEEAGPPSPKPLVLDAEQLLERELSFIRNAKESG 767
GSVYSLGVPLLADF + P E STS+ P S PLV DAEQ LE EL FI AKESG
Sbjct: 270 GSVYSLGVPLLADFKGTSNPILEASTSDVISPVSTDPLVFDAEQSLESELYFIHKAKESG 449
Query: 768 LVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMTYM 824
+VYLLGHGDIRARK+SWFIKKLVINYFYAFLRKNCRRG+T LSVPHSHLMQV MTYM
Sbjct: 450 VVYLLGHGDIRARKESWFIKKLVINYFYAFLRKNCRRGITTLSVPHSHLMQVSMTYM 620
>TC228579 similar to UP|PT13_ARATH (Q8LPL8) Potassium transporter 13
(AtPOT13) (AtKT5), partial (13%)
Length = 974
Score = 244 bits (622), Expect = 1e-64
Identities = 122/149 (81%), Positives = 133/149 (88%), Gaps = 2/149 (1%)
Frame = +3
Query: 678 AQERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLADFNESFMP--SFEPSTSE 735
AQE LES+G +DT+ EDEY+GSRVLIAP GSVYSLGVPLLADFN++ +P +FE STSE
Sbjct: 3 AQEXXLESEGXDDTDSEDEYSGSRVLIAPXGSVYSLGVPLLADFNDTTIPIPNFEASTSE 182
Query: 736 EAGPPSPKPLVLDAEQLLERELSFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYFY 795
E SPKP VLDAEQ LERELSFIR AKESG+VYLLGHGDIRARKDSWFIKKL+INYFY
Sbjct: 183 ETNLESPKPPVLDAEQSLERELSFIRKAKESGVVYLLGHGDIRARKDSWFIKKLIINYFY 362
Query: 796 AFLRKNCRRGVTNLSVPHSHLMQVGMTYM 824
AFLRKNCRRG+TNLSVPHSHLMQVGMTYM
Sbjct: 363 AFLRKNCRRGITNLSVPHSHLMQVGMTYM 449
>AW102184 similar to GP|9955527|emb potassium transport protein-like
{Arabidopsis thaliana}, partial (13%)
Length = 365
Score = 223 bits (568), Expect = 3e-58
Identities = 110/120 (91%), Positives = 115/120 (95%)
Frame = +2
Query: 417 CIKQSTALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIA 476
CIKQS ALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLV VC+ISSIDEIGNAYGIA
Sbjct: 2 CIKQSAALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVLVCSISSIDEIGNAYGIA 181
Query: 477 ELGVMMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFA 536
ELGVMMMTTILVTLVMLLIWQ+HII+V+SF VFLGLEL FFSSVLWS+TDGSWIILVFA
Sbjct: 182 ELGVMMMTTILVTLVMLLIWQIHIIVVLSFAVVFLGLELTFFSSVLWSVTDGSWIILVFA 361
>AW756492 similar to GP|9955527|emb| potassium transport protein-like
{Arabidopsis thaliana}, partial (9%)
Length = 439
Score = 221 bits (563), Expect = 1e-57
Identities = 109/132 (82%), Positives = 119/132 (89%)
Frame = +1
Query: 14 EDDPGSALSFDSTESRWVFQEDEDPSEIEDYDASDMRHQSMFDSEDEDNAEMKLIRTGPR 73
EDDP DSTESRWVFQEDEDPSEIED+DA+D+RHQSMFDS+DEDNAE +L+RTGPR
Sbjct: 61 EDDP------DSTESRWVFQEDEDPSEIEDFDAADLRHQSMFDSDDEDNAEQRLVRTGPR 222
Query: 74 IDSFDVEALEVPGAHTHHYEDMTTGKKIVLAFQTLGVVFGDVGTSPLYTFSVMFRKAPIN 133
IDSFDVEALEVPGAH + YED++ GK I+LAFQTLGVVFGDVGTSPLYTFSVMFRKAPI
Sbjct: 223 IDSFDVEALEVPGAHRNDYEDVSVGKGILLAFQTLGVVFGDVGTSPLYTFSVMFRKAPIK 402
Query: 134 DNEDILGALSLV 145
NEDILGALSLV
Sbjct: 403 GNEDILGALSLV 438
>TC217848 similar to UP|PT11_ARATH (O64769) Potassium transporter 11
(AtPOT11), partial (27%)
Length = 1192
Score = 200 bits (508), Expect = 2e-51
Identities = 106/281 (37%), Positives = 165/281 (57%), Gaps = 4/281 (1%)
Frame = +3
Query: 547 NYGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPA 606
+YGS +YE E+ K+S + LG +LG +R PGIGL+Y EL G+P IF HF+T LPA
Sbjct: 3 HYGSVKRYEFEMHSKVSMAWILGLGPSLGLVRVPGIGLVYTELASGVPHIFSHFITNLPA 182
Query: 607 IHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLL 666
IHS+++FV +KY+PV VP+ ERFL +R+ +++H+FRC+ARYGYKD K++ + FE+ L
Sbjct: 183 IHSVVVFVCVKYLPVYTVPEDERFLVKRIGPKNFHMFRCVARYGYKDLHKKD-EDFEKKL 359
Query: 667 MESLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLADFNESFM 726
+L F++ E+ D E ++ L+ N + +L + +S +
Sbjct: 360 FHNLFVFVKLESMMEGCSDSDDYSLYDEQTERSTQGLLNNNTNTAALNMDPTVSSVDSIV 539
Query: 727 PSFEP----STSEEAGPPSPKPLVLDAEQLLERELSFIRNAKESGLVYLLGHGDIRARKD 782
P +T + +G S V E+ F+ N +++G+V++LG+ +RAR+D
Sbjct: 540 SVSSPLHINATIQSSGHVSSHTEV--------DEVEFLNNCRDAGVVHILGNTVVRARRD 695
Query: 783 SWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMTY 823
S F KK+ ++Y YAFLRK CR +VPH L+ VG +
Sbjct: 696 SRFHKKIAVDYIYAFLRKICRENSVIFNVPHESLLNVGQVF 818
>BG839197
Length = 816
Score = 160 bits (406), Expect(2) = 2e-46
Identities = 88/197 (44%), Positives = 118/197 (59%), Gaps = 25/197 (12%)
Frame = +2
Query: 257 VNGLKVGVDAIQQDEVVMISVACLVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYN 316
++GL+ +D EVV IS+ LV LFS+Q++GTSKVG P L +WF SL G+YN
Sbjct: 2 ISGLQDQIDEFGTGEVVGISIVVLVALFSIQRFGTSKVGFMFAPILALWFFSLGAIGIYN 181
Query: 317 LVKYDSSVFRAFNPIHIYYFFARNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQ-- 374
++KYD +V RAFNP +IYYFF N AW +LGGC+LC TG+EAMFADL +FSV ++Q
Sbjct: 182 ILKYDITVLRAFNPAYIYYFFKNNGKDAWSALGGCVLCITGAEAMFADLGHFSVPAIQIA 361
Query: 375 --------PIIGLFGSSCIPYGTPC*CW*SLFF---------------FCSKLIASRTMT 411
++ G + P + S+F+ + +IAS+ M
Sbjct: 362 FTCVVFPCLLLAYMGQAAFLTKNPN-SYASVFYKSVPESLFWPMFVIATLAAMIASQAMI 538
Query: 412 TATFSCIKQSTALGCFP 428
+ATFSCIKQS ALGCFP
Sbjct: 539 SATFSCIKQSMALGCFP 589
Score = 44.7 bits (104), Expect(2) = 2e-46
Identities = 22/46 (47%), Positives = 28/46 (60%), Gaps = 9/46 (19%)
Frame = +1
Query: 425 GCFPRLKIIHTSRKFMGQIYIPVIN---------WFLLAVSLVFVC 461
G PRLKIIHTS++F+GQIYIP + WF L +L+ C
Sbjct: 577 GMLPRLKIIHTSKRFIGQIYIP*LTGF**SCA*LWFHLXSTLILQC 714
>BF067393
Length = 463
Score = 183 bits (465), Expect = 2e-46
Identities = 94/154 (61%), Positives = 112/154 (72%)
Frame = +2
Query: 550 SKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHS 609
SKL YET+ K LS DLM+EL CNLGTI+A IG LYNEL KGIP + GHFL++L ++HS
Sbjct: 2 SKLNYETDDKPMLSLDLMQELSCNLGTIQATVIGKLYNELEKGIPSMVGHFLSSLASLHS 181
Query: 610 MIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLMES 669
MII VSI YVP MVP SERFLF VCQRSYHLFRCIA +G KD ENH + LLMES
Sbjct: 182 MIICVSINYVPGPMVPHSERFLFPPVCQRSYHLFRCIANHGDKDVSSENHSTYYPLLMES 361
Query: 670 LEKFIRREAQERSLESDGDEDTELEDEYAGSRVL 703
L+ IR +AQ RS ES+ D+ + +D+ + VL
Sbjct: 362 LQNVIRLDAQYRSAESEWDDQLDYQDDLYSATVL 463
>TC222511 similar to UP|Q94KC5 (Q94KC5) Potassium transporter HAK3p
(Fragment), partial (48%)
Length = 616
Score = 171 bits (432), Expect = 2e-42
Identities = 80/190 (42%), Positives = 125/190 (65%)
Frame = +2
Query: 469 IGNAYGIAELGVMMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDG 528
+GNA G+A + VM++TT L++LV++L W +I++ + F+ F +E ++FS+ L +G
Sbjct: 17 MGNAAGLAVITVMLVTTCLMSLVIVLCWHKNILLAVCFILFFGSIEALYFSASLIKFLEG 196
Query: 529 SWIILVFAAIMFFIMFIWNYGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNE 588
+W+ + + I M++W+YG+ KYE +V+ K+ + + LG ++G + GIGL++ E
Sbjct: 197 AWVPIALSLIFLIAMYVWHYGTLKKYEFDVQNKVPINWLLSLGPSIGIVTVKGIGLIHTE 376
Query: 589 LVKGIPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIAR 648
LV GIP IF HF+T LPA H ++IF+ IK V V V ERFL RV + Y L+RCIAR
Sbjct: 377 LVSGIPAIFSHFVTNLPAFHQVVIFLCIKSVQVPHVRPEERFLVGRVGPKEYILYRCIAR 556
Query: 649 YGYKDARKEN 658
YGY D K++
Sbjct: 557 YGYHDIHKDD 586
>TC224679 similar to UP|Q94KC6 (Q94KC6) Potassium transporter HAK2p, partial
(28%)
Length = 1218
Score = 169 bits (429), Expect = 4e-42
Identities = 104/317 (32%), Positives = 167/317 (51%), Gaps = 25/317 (7%)
Frame = +2
Query: 533 LVFAAIMFFIMFIWNYGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKG 592
++ A + IM++W+Y + KYE ++ K+S D + LG +LG R PGIGL++ +L G
Sbjct: 2 VLLALFLMIIMYLWHYATIRKYEYDLHNKVSLDWLLALGPSLGIARVPGIGLVFTDLTTG 181
Query: 593 IPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYK 652
IP F F+T LPA H +++FV +K VPV VP +ER+L RV ++ +RCI RYGY+
Sbjct: 182 IPANFSRFVTNLPAYHRILVFVCVKSVPVPHVPAAERYLVGRVGPPAHRSYRCIVRYGYR 361
Query: 653 DARKENHQAFEQLLMESLEKFIRRE-AQERSLESDGDEDTELEDEYAGSRVLI------- 704
D ++ +FE L+ L FI+ + + R D+D +E + R+ +
Sbjct: 362 DVH-QDIDSFESELVARLADFIQYDWYRSRRSSMSIDDDASNSNESSSYRLTVIGTTGFT 538
Query: 705 ----------APNGSVYSLGVPLLADFNESFMPSFEPSTSE-------EAGPPSPKPLVL 747
+ + S+G P + + + EP +E E P S
Sbjct: 539 IQPGYESGGESVQQASVSVGFPTVQSVTD--VIEMEPVMTERRVRFAIEDEPESDARSET 712
Query: 748 DAEQLLERELSFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVT 807
+ ++ EL + A+E+G+ ++LGH +RA++ S +KKL +NY Y FLR+NCR
Sbjct: 713 GVQ--MQEELEDLYAAQEAGIAFILGHSHVRAKQGSSVLKKLALNYGYNFLRRNCRGPDV 886
Query: 808 NLSVPHSHLMQVGMTYM 824
L VP L++VGM Y+
Sbjct: 887 ALKVPPVSLLEVGMVYI 937
>BU764111 similar to GP|9955527|emb potassium transport protein-like
{Arabidopsis thaliana}, partial (16%)
Length = 445
Score = 163 bits (412), Expect = 3e-40
Identities = 93/150 (62%), Positives = 100/150 (66%), Gaps = 27/150 (18%)
Frame = +3
Query: 310 AGNGVYNLVKYDSSVFRAFNPIHIYYFFARNSTKAWYSLGGCLLCATGSEAMFADLCYFS 369
AG G+YNLVKYD+SV RAFNPIHIYYFF RNST AWYSLGGCLL ATGSEAMFADLCYFS
Sbjct: 3 AGIGIYNLVKYDNSVLRAFNPIHIYYFFKRNSTNAWYSLGGCLLSATGSEAMFADLCYFS 182
Query: 370 VRSVQ----------PIIGLFGS----------------SCIPYGTPC*CW*S-LFFFCS 402
VRSVQ ++G G S +P G W + L +
Sbjct: 183 VRSVQLSFVFLVLPCLLLGYLGQAAYLMENHADAGQAFFSSVPSGA---FWPTFLIANIA 353
Query: 403 KLIASRTMTTATFSCIKQSTALGCFPRLKI 432
LIASR MTTATFSCIKQSTALGCFPRLKI
Sbjct: 354 ALIASRAMTTATFSCIKQSTALGCFPRLKI 443
>BE191551
Length = 475
Score = 152 bits (383), Expect = 8e-37
Identities = 73/148 (49%), Positives = 101/148 (67%)
Frame = -3
Query: 227 KKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVACLVVLFSL 286
K I+L+L L GT MVI +G++TPA+SVLS+V G+KV + + VV+++V LV LFS+
Sbjct: 473 KNIILMLALVGTCMVIGDGILTPAISVLSAVGGIKVNHADLSNEVVVLVAVVILVGLFSM 294
Query: 287 QKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIHIYYFFARNSTKAWY 346
Q YGT KVG P + +WF + G G++N+ Y SSV +AF+P++IY + R W
Sbjct: 293 QHYGTDKVGWLFAPIVLLWFLLIGGIGIFNICXYGSSVLKAFSPLYIYRYLQREGKDGWL 114
Query: 347 SLGGCLLCATGSEAMFADLCYFSVRSVQ 374
SLGG LL TG+EA+FADL +F V SVQ
Sbjct: 113 SLGGILLSITGTEALFADLAHFPVSSVQ 30
>TC203534
Length = 1010
Score = 151 bits (382), Expect = 1e-36
Identities = 92/246 (37%), Positives = 138/246 (55%)
Frame = +1
Query: 578 RAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQ 637
R P IGLLY+ELV+GIP I H + +P+IHS+I+FVSI +PV+ V ER FR+V
Sbjct: 19 RVPXIGLLYSELVQGIPPISXHLIDNIPSIHSIIVFVSIXXIPVSRVASEERXXFRQVEP 198
Query: 638 RSYHLFRCIARYGYKDARKENHQAFEQLLMESLEKFIRREAQERSLESDGDEDTELEDEY 697
R Y + RC+ R+GY D E+ E L+++L+ F++ E LE DG E E E
Sbjct: 199 RDYRVXRCVVRHGYNDV-LEDPAEXESHLIQNLKAFVQHE--NYMLEVDGTEHASAETE- 366
Query: 698 AGSRVLIAPNGSVYSLGVPLLADFNESFMPSFEPSTSEEAGPPSPKPLVLDAEQLLEREL 757
+ GS + +P A + + S S ++ + SP P+ Q E E+
Sbjct: 367 ---MIAAVGKGSSNRI-IPDQAAASSDSIRSLGASATKSSSFISP-PI-----QGAEDEI 516
Query: 758 SFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLM 817
FI A E G+VY+L ++ A S + K+V+NY Y+F RKN R+G ++++ + L+
Sbjct: 517 KFIDKALEKGVVYMLAEAEVVAHPSSSILNKIVVNYVYSFFRKNFRQGQNSMAIQRNRLL 696
Query: 818 QVGMTY 823
+VGMTY
Sbjct: 697 KVGMTY 714
>BE801045
Length = 421
Score = 147 bits (372), Expect = 1e-35
Identities = 64/139 (46%), Positives = 96/139 (69%)
Frame = +1
Query: 508 GVFLGLELVFFSSVLWSITDGSWIILVFAAIMFFIMFIWNYGSKLKYETEVKQKLSPDLM 567
G+ L +E +FS+VL+ + G W L A IM++W+YG+ +YE E+ K+S +
Sbjct: 4 GLSLIVECTYFSAVLFKVDQGGWAPLAIAGAFLLIMYVWHYGTVKRYEFEMHSKVSMAWI 183
Query: 568 RELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQS 627
LG +LG +R PGIGL+Y EL G+P IF HF+T LPAIHS+++FV +KY+PV VP+
Sbjct: 184 LGLGPSLGLVRVPGIGLVYTELANGVPHIFSHFITNLPAIHSVVVFVCVKYLPVYTVPED 363
Query: 628 ERFLFRRVCQRSYHLFRCI 646
ERFL +R+ +++H+FRC+
Sbjct: 364 ERFLVKRIGPKNFHMFRCV 420
>TC232266 similar to UP|Q94KC5 (Q94KC5) Potassium transporter HAK3p
(Fragment), partial (45%)
Length = 873
Score = 145 bits (367), Expect = 5e-35
Identities = 89/258 (34%), Positives = 140/258 (53%), Gaps = 8/258 (3%)
Frame = +1
Query: 574 LGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFR 633
LG +R G+GL++ ELV GIP IF HF+T LPA H +++F+ IK+VPV V ERFL
Sbjct: 25 LGIVRVRGVGLIHTELVSGIPVIFSHFVTNLPAFHQVLVFLCIKHVPVPHVTPEERFLVG 204
Query: 634 RVCQRSYHLFRCIARYGYKDARKENHQAFEQLLMESLEKFIRREAQERSLESD---GDED 690
RV + + L+RCI RYGY+D +++ + FE L+ + +FIR E + +D D+
Sbjct: 205 RVGPKEFRLYRCIVRYGYRDVHRDDVE-FENDLLCCIAEFIRTERTGSNSSNDEPVKDDR 381
Query: 691 TELEDEYAGSRVLIAPN--GSVYSLGVPLLADFNESFMPSFEPSTSEEAG---PPSPKPL 745
+ + +L+ N +V ++ +P ++ E P+ + P SPK
Sbjct: 382 MAVVGTCSTHSLLMTENKVDNVENVDLPGPSELKEIKSPNVIQQQKKRVRFLVPESPK-- 555
Query: 746 VLDAEQLLERELSFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRG 805
+ + EL + A E+G+ Y++G +RA+ S +KK+ IN Y FLR+N R
Sbjct: 556 ---IDTSVMEELEEVMEAWEAGVAYIIGQTHMRAKSGSSMLKKIGINLVYEFLRRNSRAP 726
Query: 806 VTNLSVPHSHLMQVGMTY 823
VPH+ ++VGM Y
Sbjct: 727 SFVTGVPHASSLEVGMMY 780
>TC211592 similar to UP|POT6_ARATH (Q8W4I4) Potassium transporter 6 (AtPOT6)
(AtHAK6), partial (22%)
Length = 613
Score = 144 bits (364), Expect = 1e-34
Identities = 75/198 (37%), Positives = 120/198 (59%)
Frame = +3
Query: 516 VFFSSVLWSITDGSWIILVFAAIMFFIMFIWNYGSKLKYETEVKQKLSPDLMRELGCNLG 575
+FFS+ L G+W+ + A ++ +M+ W+YG+ KYE +V+ K+S + + + G +LG
Sbjct: 3 LFFSASLIKFLQGAWVPIALALVLLTVMYAWHYGTLKKYEYDVQNKVSINWLLDQGPSLG 182
Query: 576 TIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRV 635
+R G+GLL+ ELV GIP IF F+ LPA H +++F+ IK+VPV V ERFL R+
Sbjct: 183 IVRVHGVGLLHTELVSGIPVIFFQFVANLPAFHQVLVFLCIKHVPVPHVKAKERFLVGRI 362
Query: 636 CQRSYHLFRCIARYGYKDARKENHQAFEQLLMESLEKFIRREAQERSLESDGDEDTELED 695
+ + ++RCI RYGY D +++ + FE L+ S+ +FIR E ES+ D L+D
Sbjct: 363 GPKEFRIYRCIVRYGYHDVHRDDFE-FENDLICSIAEFIRTE----RTESNSPNDEPLKD 527
Query: 696 EYAGSRVLIAPNGSVYSL 713
+ R+ + S +SL
Sbjct: 528 D----RMAVVGTCSTHSL 569
>BF009653 similar to GP|9955527|emb| potassium transport protein-like
{Arabidopsis thaliana}, partial (8%)
Length = 222
Score = 130 bits (328), Expect = 2e-30
Identities = 65/73 (89%), Positives = 70/73 (95%)
Frame = +3
Query: 442 QIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGVMMMTTILVTLVMLLIWQMHII 501
QIYIPVINWFLLA+SLV VCTISSIDEIGNAYGIAELGVMMMTTIL TLVMLLIWQ+HII
Sbjct: 3 QIYIPVINWFLLALSLVLVCTISSIDEIGNAYGIAELGVMMMTTILATLVMLLIWQIHII 182
Query: 502 IVMSFLGVFLGLE 514
I++SF+ VFLGLE
Sbjct: 183 ILLSFVVVFLGLE 221
>CA800288
Length = 435
Score = 126 bits (316), Expect = 4e-29
Identities = 70/146 (47%), Positives = 95/146 (64%)
Frame = +1
Query: 171 GTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKERLESSFTLKKIL 230
GTFALYSL+CR+AK NLLPNQ +D +S ++ PS++ S LK LE L+ L
Sbjct: 1 GTFALYSLLCRHAKFNLLPNQQAADEELSSYKYG-PSSQAIASSPLKRFLEKHKRLRTAL 177
Query: 231 LLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVACLVVLFSLQKYG 290
L++VL G MVI +GV+TPA+SVL+SV+GLKV + E+V+++ LV LF+LQ G
Sbjct: 178 LVVVLFGACMVIGDGVLTPAISVLASVSGLKVTEKKLTDGELVLLACVILVGLFALQHCG 357
Query: 291 TSKVGLAVGPALFIWFCSLAGNGVYN 316
T KV + P + IW S+ GVYN
Sbjct: 358 THKVAVMFAPIVIIWLVSIFSIGVYN 435
>BI321387
Length = 423
Score = 123 bits (309), Expect = 3e-28
Identities = 55/115 (47%), Positives = 84/115 (72%)
Frame = +3
Query: 404 LIASRTMTTATFSCIKQSTALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTI 463
++AS+ +ATFS IKQ+ A GCFPR+K++HTS+KF+GQIYIP INW L+ + +
Sbjct: 78 IVASQATISATFSIIKQANAHGCFPRIKVVHTSKKFLGQIYIPDINWILMILCIAVTAGF 257
Query: 464 SSIDEIGNAYGIAELGVMMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFF 518
+ +IGNAYG A + VM++TT+L+ L+M+L+W+ H I+V+ F G+ L +E +F
Sbjct: 258 KNQSQIGNAYGTAVVLVMLVTTLLMILIMILVWRCHWILVVVFTGLSLIVECTYF 422
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,087,612
Number of Sequences: 63676
Number of extensions: 565960
Number of successful extensions: 4376
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 4284
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4353
length of query: 824
length of database: 12,639,632
effective HSP length: 105
effective length of query: 719
effective length of database: 5,953,652
effective search space: 4280675788
effective search space used: 4280675788
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144459.2


