

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144404.22 + phase: 0
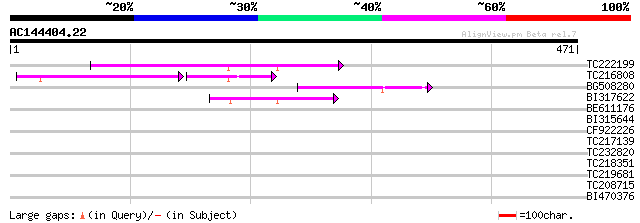
(471 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222199 97 1e-20
TC216808 56 2e-15
BG508280 69 6e-12
BI317622 55 5e-08
BE611176 34 0.16
BI315644 32 0.61
CF922226 32 0.61
TC217139 weakly similar to UP|Q6K6I0 (Q6K6I0) Nucellin-like aspa... 30 2.3
TC232820 similar to UP|Q6HFY8 (Q6HFY8) Possible transcriptional ... 30 2.3
TC218351 28 6.8
TC219681 weakly similar to UP|Q9FG60 (Q9FG60) Aspartyl protease-... 28 6.8
TC208715 28 6.8
BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sat... 28 8.8
>TC222199
Length = 691
Score = 97.4 bits (241), Expect = 1e-20
Identities = 62/226 (27%), Positives = 107/226 (46%), Gaps = 16/226 (7%)
Frame = +2
Query: 68 DYQILPTLEEYSCWINLPVLDKVPFSGLEETPKHSTIAATLHLETDEVKTHLITRGKFLG 127
D+Q++PT+EE+ + P+ ++ P+ P S I + + TR G
Sbjct: 5 DFQLVPTIEEFEEILGCPLGERKPYLSSGCLPSLSRIPTMVKDSARGLDRVKQTRNGIAG 184
Query: 128 FSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFVDIKAIQIFLS-----KN 182
+L ++ G F +LALL++G+VLFP++D VD+ AI FL+ ++
Sbjct: 185 LPRKYLEDKARGIANQGDWVPFMDVLALLIFGVVLFPNVDGLVDLAAIDAFLAYHHSKES 364
Query: 183 PVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLL---------PRTPHFT-TNPE 232
PV +L D + + +RR + I+CC L W+ S L P H + T
Sbjct: 365 PVVAVLADLFDTFNRRYEKSSARIICCLPALCVWLVSHLFQQDTRRPCPLLSHRSCTEKR 544
Query: 233 NLLWSKRLMSLTPAEVVWYDRVYD-KGTIIDSCGKFANVPLLGMEG 277
+ W + L + + W+ R + K ++ SCG++ N+PL+G G
Sbjct: 545 RIDWDQLLAGIGGRTISWFPRWKEGKEGVLFSCGRYPNIPLVGTRG 682
>TC216808
Length = 1144
Score = 56.2 bits (134), Expect(2) = 2e-15
Identities = 31/79 (39%), Positives = 47/79 (59%), Gaps = 5/79 (6%)
Frame = +3
Query: 148 AFNSILALLVYGLVLFPSLDNFVDIKAIQIFLS-----KNPVPTLLGDTYLSIHRRTQAG 202
+F ILALL++G+VLFP+++ VD+ AI FL+ ++PV +L Y + R +
Sbjct: 591 SFIDILALLIFGVVLFPNMEGLVDLAAIDAFLAFHHGKESPVVAILA-MYDTFDRGCEKS 767
Query: 203 RGTILCCAQLLYRWITSLL 221
I+CCA LY W+ S L
Sbjct: 768 SARIVCCALALYVWLVSHL 824
Score = 44.3 bits (103), Expect(2) = 2e-15
Identities = 30/141 (21%), Positives = 62/141 (43%), Gaps = 2/141 (1%)
Frame = +2
Query: 6 QYSFRKPDLLSLRKLGKKV--LCTDDFFEQHGNLLGVVKTDIDEGLLNAFVQFYDPGYHC 63
Q + ++ SL +LG+ + L F + +G + + + ++ + + Q+YD C
Sbjct: 158 QVKIKSLEVTSLGELGQLMGQLQR*AFRKAYGKIWDLARVEVSMEPIASLSQYYDQPLRC 337
Query: 64 FTFQDYQILPTLEEYSCWINLPVLDKVPFSGLEETPKHSTIAATLHLETDEVKTHLITRG 123
FTF+D+Q++PT++E + + P+ P + +A + + E+ R
Sbjct: 338 FTFEDFQLVPTVKELEEIRGCQLGGRKPYLFSGFYPSTTRVAKVVKISAQELNRVKQNRN 517
Query: 124 KFLGFSTDFLYERTTFFDKMG 144
+G L ER + G
Sbjct: 518 GVVGVPRKCLEERAKTLENQG 580
>BG508280
Length = 418
Score = 68.6 bits (166), Expect = 6e-12
Identities = 44/115 (38%), Positives = 62/115 (53%), Gaps = 3/115 (2%)
Frame = +1
Query: 240 LMSLTPAEVVWYDRVYDKGT-IIDSCGKFANVPLLGMEGGISYNPTLARRQFGYPMEKKP 298
L S A V W+ R + T ++ SCG F NVPL+G G ISYNP LA +Q GYPM P
Sbjct: 7 LASKEGASVNWFPRWKEGRTRVLISCGGFPNVPLMGTRGCISYNPVLAIKQLGYPM*GVP 186
Query: 299 LSIYLENVYY--FNTEDSTGMREQVVRAWHTIRRRDKGQLGKKTGAVHESYTQWV 351
L L V FN + + ++V +AW ++++DK G G + Y +W+
Sbjct: 187 LEDELAPVISRGFN-KTNVETLQKVRKAWEVLQKKDKELRGSNNGPI-GGYRKWL 345
>BI317622
Length = 370
Score = 55.5 bits (132), Expect = 5e-08
Identities = 39/123 (31%), Positives = 57/123 (45%), Gaps = 16/123 (13%)
Frame = -2
Query: 167 DNFVDIKAIQIFLSKN-----PVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLL 221
D VD+ AI FL+ + P+ +L D Y + +RR + I+CC LY W+ S L
Sbjct: 369 DGLVDLAAIGAFLAYHDYKEIPIVAMLADLYDTFNRRCEKSSTRIVCCTPALYVWLVSHL 190
Query: 222 ---------PRTPHFT-TNPENLLWSKRLMSLTPAEVVWYDRVYDKGT-IIDSCGKFANV 270
P H + T W + L + A V W+ R + T ++ SCG F NV
Sbjct: 189 FRQEVRHACPLESHCSCTEKGGENWGQLLANKEGASVNWFPRWKEGRTRVLISCGVFLNV 10
Query: 271 PLL 273
PL+
Sbjct: 9 PLM 1
>BE611176
Length = 424
Score = 33.9 bits (76), Expect = 0.16
Identities = 18/68 (26%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Frame = -2
Query: 6 QYSFRKPDLLSLRKLGKKV--LCTDDFFEQHGNLLGVVKTDIDEGLLNAFVQFYDPGYHC 63
Q + D+ S+++LG+ + L F + + +L + +I + + Q+YD C
Sbjct: 348 QVKIKGLDITSIKELGRLMGPLQMQAFRKAYEKILDLTIAEISTEAIASLTQYYD*PLRC 169
Query: 64 FTFQDYQI 71
FTF D+Q+
Sbjct: 168 FTFGDFQL 145
>BI315644
Length = 421
Score = 32.0 bits (71), Expect = 0.61
Identities = 14/22 (63%), Positives = 15/22 (67%)
Frame = +3
Query: 277 GGISYNPTLARRQFGYPMEKKP 298
G I+YNP LA RQ GYPM P
Sbjct: 282 GCINYNPVLAIRQLGYPMRGVP 347
>CF922226
Length = 667
Score = 32.0 bits (71), Expect = 0.61
Identities = 25/72 (34%), Positives = 35/72 (47%), Gaps = 1/72 (1%)
Frame = -3
Query: 386 ECQEKIAELTRESYMWERKYRKKEQEYDTLKNLLDQQIQANRQEKEENI-RLRAALQKKE 444
E +EK + + E K KK+ E+D K Q N++ E NI ++R KKE
Sbjct: 386 ERKEKKSSASGEGLTARGKTFKKDSEFDKKK-----QKPENQKNGEGNIFKIRCYHCKKE 222
Query: 445 DFLDKVCPGRKK 456
KVCP R+K
Sbjct: 221 GHTRKVCPERQK 186
>TC217139 weakly similar to UP|Q6K6I0 (Q6K6I0) Nucellin-like aspartic
protease-like, partial (61%)
Length = 967
Score = 30.0 bits (66), Expect = 2.3
Identities = 20/55 (36%), Positives = 27/55 (48%)
Frame = +1
Query: 217 ITSLLPRTPHFTTNPENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVP 271
+T LLP PH++ N + +SL+ DR KGTIIDS A +P
Sbjct: 562 MTPLLPDQPHYSVNMTAVQVGHAFLSLSTDTSTQGDR---KGTIIDSGTTLAYLP 717
>TC232820 similar to UP|Q6HFY8 (Q6HFY8) Possible transcriptional regulator,
PadR family, partial (15%)
Length = 918
Score = 30.0 bits (66), Expect = 2.3
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = -2
Query: 217 ITSLLPRTPHFTTNPENLLWS 237
+ SL+PRTP FTTN L W+
Sbjct: 524 LKSLIPRTPLFTTNHVKLFWN 462
>TC218351
Length = 595
Score = 28.5 bits (62), Expect = 6.8
Identities = 14/30 (46%), Positives = 20/30 (66%)
Frame = +1
Query: 159 GLVLFPSLDNFVDIKAIQIFLSKNPVPTLL 188
G LFPS+ F+ K++ FLSKNP+ + L
Sbjct: 346 GSFLFPSMILFI-CKSLSFFLSKNPIASCL 432
>TC219681 weakly similar to UP|Q9FG60 (Q9FG60) Aspartyl protease-like,
partial (45%)
Length = 1046
Score = 28.5 bits (62), Expect = 6.8
Identities = 17/56 (30%), Positives = 31/56 (55%)
Frame = +2
Query: 218 TSLLPRTPHFTTNPENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLL 273
T L+PR H+ +++ ++ L P+++ +D V KGT+IDS A +P +
Sbjct: 341 TPLVPRMAHYNVVLKSIEVDTDILQL-PSDI--FDSVNGKGTVIDSGTTLAYLPAI 499
>TC208715
Length = 950
Score = 28.5 bits (62), Expect = 6.8
Identities = 13/46 (28%), Positives = 23/46 (49%), Gaps = 6/46 (13%)
Frame = +2
Query: 206 ILCCAQLLYRWITS------LLPRTPHFTTNPENLLWSKRLMSLTP 245
I CC L +RW+ + P + HF P++ + + L+S +P
Sbjct: 659 ISCCIWLTWRWVLTTESAPCATPNSSHFARTPKSRVENFSLLSFSP 796
>BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sativa
(japonica cultivar-group)}, partial (5%)
Length = 429
Score = 28.1 bits (61), Expect = 8.8
Identities = 12/50 (24%), Positives = 28/50 (56%)
Frame = -1
Query: 37 LLGVVKTDIDEGLLNAFVQFYDPGYHCFTFQDYQILPTLEEYSCWINLPV 86
++ + + ++ L+NAF++ + P H F + + TL++ S + +PV
Sbjct: 369 IMKLAQLKVNGALVNAFIERWRPETHTFHLKYGEATITLQDVSVLLGIPV 220
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,341,371
Number of Sequences: 63676
Number of extensions: 305508
Number of successful extensions: 1533
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1518
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1527
length of query: 471
length of database: 12,639,632
effective HSP length: 101
effective length of query: 370
effective length of database: 6,208,356
effective search space: 2297091720
effective search space used: 2297091720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144404.22


