

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144389.8 - phase: 0
(468 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
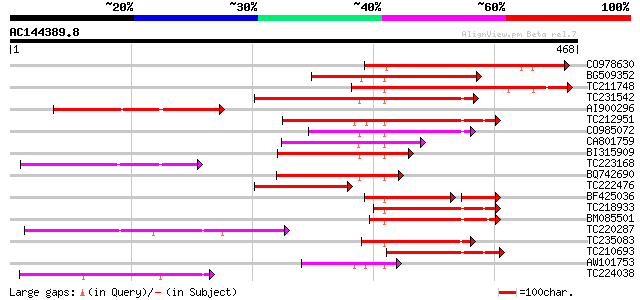
Score E
Sequences producing significant alignments: (bits) Value
CO978630 244 8e-65
BG509352 weakly similar to GP|18700084|gb At2g46620/F13A10.15 {A... 197 1e-50
TC211748 similar to GB|AAN28905.1|23506091|AY143966 At2g46620/F1... 196 1e-50
TC231542 similar to UP|Q8LBK2 (Q8LBK2) BCS1 protein-like protein... 194 7e-50
AI900296 weakly similar to GP|18700084|gb At2g46620/F13A10.15 {A... 193 1e-49
TC212951 similar to UP|Q8LBK2 (Q8LBK2) BCS1 protein-like protein... 153 1e-37
CO985072 120 1e-27
CA801759 similar to GP|9294272|dbj mitochondrial protein-like {A... 114 7e-26
BI315909 similar to GP|18086343|gb AT3g50930/F18B3_210 {Arabidop... 113 2e-25
TC223168 similar to UP|Q8V7E3 (Q8V7E3) ORF1 (Fragment), partial ... 110 1e-24
BQ742690 similar to GP|9294272|dbj mitochondrial protein-like {A... 107 1e-23
TC222476 similar to UP|Q9FN75 (Q9FN75) AAA-type ATPase-like prot... 103 2e-22
BF425036 weakly similar to GP|9294274|dbj| mitochondrial protein... 88 5e-21
TC218933 similar to UP|Q9FN78 (Q9FN78) AAA-type ATPase-like prot... 97 1e-20
BM085501 similar to GP|9759053|dbj AAA-type ATPase-like protein ... 96 3e-20
TC220287 similar to UP|Q9FRW0 (Q9FRW0) Mitochondrial protein-lik... 91 8e-19
TC235083 similar to UP|Q9LH82 (Q9LH82) Similarity to AAA-type AT... 91 8e-19
TC210693 weakly similar to UP|Q9FN78 (Q9FN78) AAA-type ATPase-li... 81 9e-16
AW101753 79 3e-15
TC224038 weakly similar to UP|Q9LCV5 (Q9LCV5) Atp operon (Fragme... 66 3e-11
>CO978630
Length = 660
Score = 244 bits (622), Expect = 8e-65
Identities = 130/181 (71%), Positives = 149/181 (81%), Gaps = 12/181 (6%)
Frame = -1
Query: 294 VTSSGILNFMDGIWSG---EERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKT 350
+++SGILNFMD + + EERVMVFTMN+KE+VDPNLLRPGRVDVHIHFPLCDFS+FKT
Sbjct: 654 ISASGILNFMDALLTSCCAEERVMVFTMNTKEHVDPNLLRPGRVDVHIHFPLCDFSAFKT 475
Query: 351 LASNYLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVITALKTDGDG 410
LAS+YLGVK+HKLFPQVQEIF+NGASLSPAEIGELMIANRNSPSRAIK+VITAL+TDGDG
Sbjct: 474 LASSYLGVKEHKLFPQVQEIFQNGASLSPAEIGELMIANRNSPSRAIKSVITALQTDGDG 295
Query: 411 RGCGFIERRIG----NEGDGVDEG-----ARDTRKLYGFFRLKGPRKSSSSPNSVLVASP 461
RGCG I R+ +E DGV G +D RKLYGFF + R+SSSS S+ ASP
Sbjct: 294 RGCGLIGRQTEDDEMDEPDGVVCGETFHTVKDLRKLYGFFSFRVTRRSSSSNASLPPASP 115
Query: 462 L 462
L
Sbjct: 114 L 112
>BG509352 weakly similar to GP|18700084|gb At2g46620/F13A10.15 {Arabidopsis
thaliana}, partial (30%)
Length = 436
Score = 197 bits (500), Expect = 1e-50
Identities = 103/145 (71%), Positives = 122/145 (84%), Gaps = 5/145 (3%)
Frame = +1
Query: 250 VYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYLTEK--SSTTVTSSGILNFMDGIW 307
VY +DLS+I DSDL +L +T KS+IVVEDLDR+L + ++T VT+SGI +FMDGI
Sbjct: 1 VYDVDLSKIRGDSDLMFLLTETTAKSVIVVEDLDRFLEPEPETATAVTASGIQSFMDGIV 180
Query: 308 S---GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLF 364
S GEERVMVFTMNSKE VDP+LLRPGRVDVHIHFP+CDFS+FKTLAS+YLGV+ HKLF
Sbjct: 181 SACCGEERVMVFTMNSKECVDPDLLRPGRVDVHIHFPVCDFSAFKTLASSYLGVRKHKLF 360
Query: 365 PQVQEIFENGASLSPAEIGELMIAN 389
QV+EIF +GA+LSPAEI ELMIAN
Sbjct: 361 AQVEEIFRHGATLSPAEISELMIAN 435
>TC211748 similar to GB|AAN28905.1|23506091|AY143966 At2g46620/F13A10.15
{Arabidopsis thaliana;} , partial (22%)
Length = 786
Score = 196 bits (499), Expect = 1e-50
Identities = 112/198 (56%), Positives = 137/198 (68%), Gaps = 16/198 (8%)
Frame = +2
Query: 283 DRYLTEKSSTTVTS-SGILNFMDGIWS--GEERVMVFTMN-SKENVDPNLLRPGRVDVHI 338
DR LTEKS + TS S +LNFMDGI S GEERVMVFTMN +KE VD +LRPGR+DVHI
Sbjct: 2 DRLLTEKSKSNTTSLSSVLNFMDGIVSCCGEERVMVFTMNETKEEVDQAVLRPGRIDVHI 181
Query: 339 HFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIK 398
HFPLCDFS+FK LAS+YLG+K+HKLFPQV+E+F+ GA LSPAE+GE+MI+NRNSP+RA+K
Sbjct: 182 HFPLCDFSTFKILASSYLGLKEHKLFPQVEEVFQTGARLSPAELGEIMISNRNSPTRALK 361
Query: 399 TVITALKTDGDG--RGCGFIERRIGNEGDGVDEGA----------RDTRKLYGFFRLKGP 446
TVI+AL+ +G G G D + GA R+ RKLYG RL G
Sbjct: 362 TVISALQVQSNGPREGQRLSHSGSGRNSDDNEPGAVICRESVHTVREFRKLYGLLRL-GS 538
Query: 447 RKSSSSPNSVLVASPLRI 464
R+ S + + P RI
Sbjct: 539 RRKEESYSGPIEKEPPRI 592
>TC231542 similar to UP|Q8LBK2 (Q8LBK2) BCS1 protein-like protein, partial
(34%)
Length = 1025
Score = 194 bits (493), Expect = 7e-50
Identities = 95/200 (47%), Positives = 138/200 (68%), Gaps = 15/200 (7%)
Frame = +1
Query: 203 DLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDS 262
DLE F+K K+YY R+G+ WKR +LLYG GTGKSS IAAMAN+L +DVY ++L+ ++ +S
Sbjct: 1 DLERFVKRKEYYRRVGKAWKRGYLLYGPPGTGKSSLIAAMANYLKFDVYDLELTELNANS 180
Query: 263 DLKSILLQTAPKSIIVVEDLDRYLT-------------EKSSTTVTSSGILNFMDGIWS- 308
+L+ +L+ A +SI+VVED+D + + VT SG+LNF+DG+WS
Sbjct: 181 ELRRLLIAMANRSILVVEDIDCTVEFHDRRAEARAASGHNNDRQVTLSGLLNFIDGLWSS 360
Query: 309 -GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQV 367
G+ER++VFT N K+ +DP LLRPGR+DVHIH C F+ LASNYLG+K+H LF ++
Sbjct: 361 CGDERIIVFTTNHKDKLDPALLRPGRMDVHIHMSYCTPCGFRQLASNYLGIKEHSLFEKI 540
Query: 368 QEIFENGASLSPAEIGELMI 387
+E + ++PAE+ E ++
Sbjct: 541 EEEMQK-TQVTPAEVAEQLL 597
>AI900296 weakly similar to GP|18700084|gb At2g46620/F13A10.15 {Arabidopsis
thaliana}, partial (23%)
Length = 420
Score = 193 bits (491), Expect = 1e-49
Identities = 95/142 (66%), Positives = 110/142 (76%), Gaps = 1/142 (0%)
Frame = +3
Query: 37 KLWRIIEDWFHVYQVFHVPELNDNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQN 96
KLWR IEDWFHVYQ F VPELND QHN LYRK+SLY HSLPS+++S NL+T QN
Sbjct: 6 KLWRRIEDWFHVYQFFKVPELNDTTQHNHLYRKVSLYLHSLPSIEDSDFANLITGK-KQN 182
Query: 97 DVVLTLAPNQTIHDHFLGATVSWFNQTQPNRTFILRIRKFDKQRILRAYIQHIHAVVDEI 156
D+VL L PNQTI DHFLGAT+ WFNQT TF+L+IRK DK+RILR Y+QHIHAV DEI
Sbjct: 183 DIVLCLGPNQTIEDHFLGATLFWFNQT---GTFLLKIRKVDKRRILRPYLQHIHAVADEI 353
Query: 157 EKQGNRDLRFYMN-ASDFGPWR 177
+++G RDL +MN A DF WR
Sbjct: 354 DQRGKRDLLLFMNIADDFRRWR 419
>TC212951 similar to UP|Q8LBK2 (Q8LBK2) BCS1 protein-like protein, partial
(26%)
Length = 808
Score = 153 bits (387), Expect = 1e-37
Identities = 85/204 (41%), Positives = 127/204 (61%), Gaps = 24/204 (11%)
Frame = +1
Query: 226 LLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLD-- 283
LLYG GTGKSS IAA+AN+L DVY ++LS + ++S+L ++ +T +SIIV+ED+D
Sbjct: 1 LLYGPXGTGKSSLIAAIANYLKXDVYDLELSSMXSNSELMRVMRETTXRSIIVIEDIDCN 180
Query: 284 -----RYLTEKSSTT---------------VTSSGILNFMDGIWS--GEERVMVFTMNSK 321
R T+ S + T SG+LN MDG+WS GEER+++FT N +
Sbjct: 181 KEVHARPTTKPFSDSDSDFDRKRVKVKPYRFTLSGLLNNMDGLWSSGGEERIIIFTTNHR 360
Query: 322 ENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGASLSPAE 381
E +DP LLRPGR+D+HIH +F+ LASNYLG++DH LF ++ + E ++PA
Sbjct: 361 ERIDPALLRPGRMDMHIHLSFLKGKAFRVLASNYLGIEDHSLFEEIDGLLEK-LEVTPAV 537
Query: 382 IGELMIANRNSPSRAIKTVITALK 405
+ E ++ N + P A++ ++ LK
Sbjct: 538 VAEQLMRNED-PEVALEGLVEFLK 606
>CO985072
Length = 839
Score = 120 bits (302), Expect = 1e-27
Identities = 64/170 (37%), Positives = 99/170 (57%), Gaps = 32/170 (18%)
Frame = -3
Query: 247 SYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYLT------------------- 287
+YDVY ++L+ + +SDL+ +L+ T+ KSI+V+ED+D L
Sbjct: 837 NYDVYDLELTAVKDNSDLRKLLINTSSKSIMVIEDIDCSLDLTGQRKKRKEKVEGREGKD 658
Query: 288 -----------EKSSTTVTSSGILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRV 334
+ + VT SG+LN +DGIWS G ER+MVFT N E +DP L+R GR+
Sbjct: 657 SRKRGDEDDDDDDRGSKVTLSGLLNVIDGIWSACGGERIMVFTTNFVEKLDPALIRRGRM 478
Query: 335 DVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGASLSPAEIGE 384
D HI C + +FK LA NYLG++ H+LFP+++++ E ++PA++ E
Sbjct: 477 DKHIELSYCCYEAFKVLAQNYLGLESHQLFPKIEKLLEE-TKMTPADVAE 331
>CA801759 similar to GP|9294272|dbj mitochondrial protein-like {Arabidopsis
thaliana}, partial (26%)
Length = 450
Score = 114 bits (286), Expect = 7e-26
Identities = 62/148 (41%), Positives = 88/148 (58%), Gaps = 29/148 (19%)
Frame = +3
Query: 225 FLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDR 284
+LLYG GTGKS+ IAA+ANF++YDVY ++L+ + +++L+ +L++T KSI V+ED+D
Sbjct: 3 YLLYGPPGTGKSTMIAAIANFMNYDVYDLELTAVKDNTELRKLLIETPSKSITVIEDIDC 182
Query: 285 YL---------------------------TEKSSTTVTSSGILNFMDGIWS--GEERVMV 315
L S+ VT SG+LNF+DGIWS G ER++V
Sbjct: 183 SLDLTGQRKKKKEENEDEEQKDPMRRNEEESSKSSKVTLSGLLNFIDGIWSACGGERIIV 362
Query: 316 FTMNSKENVDPNLLRPGRVDVHIHFPLC 343
FT N E +DP L+R GR+D HI C
Sbjct: 363 FTTNYVEKLDPALIRRGRMDKHIEMSYC 446
>BI315909 similar to GP|18086343|gb AT3g50930/F18B3_210 {Arabidopsis
thaliana}, partial (19%)
Length = 417
Score = 113 bits (282), Expect = 2e-25
Identities = 62/138 (44%), Positives = 85/138 (60%), Gaps = 26/138 (18%)
Frame = +2
Query: 222 KRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVED 281
KR +LL G GTGKSS IAAMAN+L++DVY ++L+ + ++DL+ +L+ T +SI+VVED
Sbjct: 2 KRGYLLSGPPGTGKSSLIAAMANYLNFDVYDLELTDVRRNTDLRKLLIGTGNRSILVVED 181
Query: 282 LDRYLT------------------------EKSSTTVTSSGILNFMDGIWS--GEERVMV 315
+D LT + VT SG LNF+DG+WS G+ER++V
Sbjct: 182 IDCSLTLQDRLAKPKSSQPVAITPWPFHPHDNPKPQVTLSGFLNFIDGLWSSCGDERIIV 361
Query: 316 FTMNSKENVDPNLLRPGR 333
FT N K +DP LLRPGR
Sbjct: 362 FTTNHKNKLDPALLRPGR 415
>TC223168 similar to UP|Q8V7E3 (Q8V7E3) ORF1 (Fragment), partial (38%)
Length = 673
Score = 110 bits (275), Expect = 1e-24
Identities = 60/151 (39%), Positives = 90/151 (58%), Gaps = 1/151 (0%)
Frame = +1
Query: 10 LLKIAILLFSIYAIHMIF-ETGLIHESTKLWRIIEDWFHVYQVFHVPELNDNMQHNTLYR 68
+L + LFS + I F +T +H + + E+ H++Q F +P N + N+LYR
Sbjct: 193 MLSLFFFLFSSFLIVFFFRKTSALHILNQWFLSFENRLHLHQSFKIPRYNLHSLDNSLYR 372
Query: 69 KLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVSWFNQTQPNRT 128
K+ Y SLPS+++S NL S N +D+ L L PN T+HD FLGA +SW N +
Sbjct: 373 KILTYLDSLPSVEDSDYTNLF-SGPNPSDIFLHLDPNHTVHDTFLGARLSWTNAS--GDA 543
Query: 129 FILRIRKFDKQRILRAYIQHIHAVVDEIEKQ 159
+LR++K DK+R+ R Y QHI +V DEIE++
Sbjct: 544 LVLRLKKKDKRRVFRQYFQHILSVADEIEQR 636
>BQ742690 similar to GP|9294272|dbj mitochondrial protein-like {Arabidopsis
thaliana}, partial (24%)
Length = 405
Score = 107 bits (266), Expect = 1e-23
Identities = 58/134 (43%), Positives = 82/134 (60%), Gaps = 29/134 (21%)
Frame = +2
Query: 221 WKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVE 280
WKR +LLYG GTGKS+ IAAMANFL YD+Y ++L+ + +++L+ +L++T+ KSIIV+E
Sbjct: 2 WKRGYLLYGPPGTGKSTMIAAMANFLGYDLYDLELTAVKDNTELRKLLIETSSKSIIVIE 181
Query: 281 DLDRYLT---------------------------EKSSTTVTSSGILNFMDGIWS--GEE 311
D+D L E S+ VT SG+LNF+DG+WS G E
Sbjct: 182 DIDCSLDLTGQRRKKKEEVEEKDQRQKQQGMQEREVKSSQVTLSGLLNFIDGLWSACGGE 361
Query: 312 RVMVFTMNSKENVD 325
R++VFT N E +D
Sbjct: 362 RLIVFTTNYVEKLD 403
>TC222476 similar to UP|Q9FN75 (Q9FN75) AAA-type ATPase-like protein, partial
(18%)
Length = 480
Score = 103 bits (257), Expect = 2e-22
Identities = 47/81 (58%), Positives = 64/81 (78%)
Frame = +1
Query: 203 DLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDS 262
DL+ F+K K++Y R+GR WKR +LLYG GTGKSS IAAMAN+L +DV+ ++L I DS
Sbjct: 22 DLDRFVKRKEFYKRVGRAWKRGYLLYGPPGTGKSSLIAAMANYLKFDVFDLELGSIVRDS 201
Query: 263 DLKSILLQTAPKSIIVVEDLD 283
DL+ +LL TA +SI+V+ED+D
Sbjct: 202 DLRKLLLATANRSILVIEDID 264
>BF425036 weakly similar to GP|9294274|dbj| mitochondrial protein-like
{Arabidopsis thaliana}, partial (15%)
Length = 416
Score = 87.8 bits (216), Expect(2) = 5e-21
Identities = 41/77 (53%), Positives = 56/77 (72%), Gaps = 2/77 (2%)
Frame = -2
Query: 294 VTSSGILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTL 351
VT SG+LNF DG+WS GEER++VFT N +++VDP L+R GR+DVH+ C +F+ L
Sbjct: 412 VTLSGLLNFTDGLWSCCGEERIVVFTTNHRDSVDPALVRCGRMDVHVSLATCGAHAFREL 233
Query: 352 ASNYLGVKDHKLFPQVQ 368
A NYLG++ H LF V+
Sbjct: 232 ARNYLGLESHVLFQAVE 182
Score = 31.6 bits (70), Expect(2) = 5e-21
Identities = 11/32 (34%), Positives = 23/32 (71%)
Frame = -3
Query: 374 GASLSPAEIGELMIANRNSPSRAIKTVITALK 405
G +L+PA++GE+++ NR A++ V+ A++
Sbjct: 165 GGALTPAQVGEILLRNRGDADVAMREVLAAMQ 70
>TC218933 similar to UP|Q9FN78 (Q9FN78) AAA-type ATPase-like protein, partial
(21%)
Length = 803
Score = 97.4 bits (241), Expect = 1e-20
Identities = 48/108 (44%), Positives = 72/108 (66%), Gaps = 3/108 (2%)
Frame = +3
Query: 301 NFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGV 358
NF+DG+WS G+ER+++FT N KE +DP LLRPGR+D+HIH C + FK LASNYL
Sbjct: 3 NFIDGLWSSCGDERIIIFTTNHKERLDPALLRPGRMDMHIHMSYCSYQGFKILASNYLET 182
Query: 359 -KDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVITALK 405
DH LF +V+ + E+ ++PA++ E ++ N + P ++ + LK
Sbjct: 183 PSDHPLFGEVEGLIED-IQITPAQVAEELMKNED-PEATLEGFVKLLK 320
>BM085501 similar to GP|9759053|dbj AAA-type ATPase-like protein {Arabidopsis
thaliana}, partial (21%)
Length = 421
Score = 96.3 bits (238), Expect = 3e-20
Identities = 46/111 (41%), Positives = 77/111 (68%), Gaps = 3/111 (2%)
Frame = +3
Query: 298 GILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNY 355
G+LNF+DG+WS G+ER+++ T N KE +DP LLRPGR+D+HIH C + FK LASNY
Sbjct: 42 GLLNFIDGLWSSCGDERIIILTTNHKERLDPALLRPGRMDMHIHMSYCSYHGFKVLASNY 221
Query: 356 LGV-KDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVITALK 405
L + DH+L +++ + E+ ++PA++ E ++ + ++ + A++ + LK
Sbjct: 222 LDIAPDHRLVGEIEGLIED-MQITPAQVAEELMKSEDADT-ALEGFLKLLK 368
>TC220287 similar to UP|Q9FRW0 (Q9FRW0) Mitochondrial protein-like protein
(Fragment), partial (44%)
Length = 980
Score = 91.3 bits (225), Expect = 8e-19
Identities = 64/244 (26%), Positives = 108/244 (44%), Gaps = 25/244 (10%)
Frame = +3
Query: 13 IAILLFSIYAIHMIFETGLIHESTKLWRIIEDWFHVYQVFHVPELN-DNMQHNTLYRKLS 71
+A ++F F L + + + + F+ Y PE + + ++ + Y +
Sbjct: 240 MATIVFMYTIFERFFPPHLREKLQAYTQKLTNHFNPYIQISFPEFSGERLKKSEAYTAIQ 419
Query: 72 LYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATV-------------- 117
Y + S + +L V N +Q +VL++ N+ I D F G +
Sbjct: 420 TYLSANSSQRAKRLKAEVV-NDSQTPLVLSMDDNEEITDEFHGIKLWRSANKVSNNPQRY 596
Query: 118 ---SWFNQTQPNRTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFG 174
S++ + R + L K + + +YI+H+ +IE + NR L+ Y N G
Sbjct: 597 NPFSYYGSSDEKRFYKLTFHKRHRDIVTMSYIKHVLDEGKDIEMR-NRQLKLYTNNPSSG 773
Query: 175 -------PWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLL 227
W + F HP+TFET+ M+ K + DL F KGK YY ++G+ WKR +LL
Sbjct: 774 WYGYKQSKWSHIVFEHPATFETLAMDRRKKEDILKDLVKFKKGKDYYAKIGKAWKRGYLL 953
Query: 228 YGSS 231
YG S
Sbjct: 954 YGPS 965
>TC235083 similar to UP|Q9LH82 (Q9LH82) Similarity to AAA-type ATPase,
partial (24%)
Length = 724
Score = 91.3 bits (225), Expect = 8e-19
Identities = 45/96 (46%), Positives = 64/96 (65%), Gaps = 2/96 (2%)
Frame = +3
Query: 291 STTVTSSGILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSF 348
++ VT SG+LNF+DGIWS G ER+++FT N + +DP L+R GR+D HI C F +F
Sbjct: 111 NSKVTLSGLLNFIDGIWSACGGERIIIFTTNFVDKLDPALIRTGRMDKHIELSYCRFEAF 290
Query: 349 KTLASNYLGVKDHKLFPQVQEIFENGASLSPAEIGE 384
K LA NYL V H LF ++ + E +++PA+I E
Sbjct: 291 KVLAKNYLDVDSHYLFARIANLLE-VTNVTPADIAE 395
>TC210693 weakly similar to UP|Q9FN78 (Q9FN78) AAA-type ATPase-like protein,
partial (18%)
Length = 520
Score = 81.3 bits (199), Expect = 9e-16
Identities = 41/97 (42%), Positives = 62/97 (63%)
Frame = +1
Query: 312 RVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIF 371
R++VFT N K +DP LLRPGR+DVHI C FK LA NYLG+ +H LF +V+ +
Sbjct: 1 RIIVFTTNHKNKLDPALLRPGRMDVHIDMTYCTPCGFKMLAFNYLGITEHPLFVEVETLL 180
Query: 372 ENGASLSPAEIGELMIANRNSPSRAIKTVITALKTDG 408
+ +++PAE+GE + N + P A+++++ L G
Sbjct: 181 KT-TNVTPAEVGEQFLKNED-PEIALESLMELLIEKG 285
>AW101753
Length = 341
Score = 79.3 bits (194), Expect = 3e-15
Identities = 46/113 (40%), Positives = 67/113 (58%), Gaps = 31/113 (27%)
Frame = +2
Query: 242 MANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLD------------------ 283
MANFL YDVY ++L+++S +S+L+S+L+QT +SIIV+ED+D
Sbjct: 2 MANFLCYDVYDLELTKVSDNSELRSLLIQTTNRSIIVIEDIDCSVDITADRTVKVKKSQG 181
Query: 284 -----RYLTEKSST------TVTSSGILNFMDGIWS--GEERVMVFTMNSKEN 323
R +K T VT SG+LNF DG+WS GEER++VFT N +++
Sbjct: 182 AKLSLRSSNKKGQTGCEESGRVTLSGLLNFTDGLWSCCGEERIVVFTTNHRDS 340
>TC224038 weakly similar to UP|Q9LCV5 (Q9LCV5) Atp operon (Fragment), partial
(4%)
Length = 559
Score = 66.2 bits (160), Expect = 3e-11
Identities = 44/171 (25%), Positives = 80/171 (46%), Gaps = 10/171 (5%)
Frame = -3
Query: 9 ILLKIAILLFSIYAIHMIFETGLIHESTKLWRIIEDWFHVYQVFHVPELND--NMQHNTL 66
IL ++ LL + + + + L+ L+ ++D Y F +PE N ++ N L
Sbjct: 548 ILSQMWSLLGLLTVLQNVLPSQLLSLLHSLYESLQDLLSPYSYFEIPEFNGYCGVELNDL 369
Query: 67 YRKLSLYFHSLPSLQNSQLNNLVTS-NTNQNDVVLTLAPNQTIHDHFLGATVSWFNQT-- 123
YR + LY ++ + L S + + N + +APN T+HD F G V W +
Sbjct: 368 YRHVHLYLNAANHAPAAACRRLTLSCSPSSNRISFAVAPNHTVHDAFRGHRVGWTHHVET 189
Query: 124 -----QPNRTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMN 169
+ R+F LR+ K + +L Y+ H+ + +E E+ +R+ R + N
Sbjct: 188 AQDSLEERRSFTLRLPKRHRHALLSPYLAHVTSRAEEFERV-SRERRLFTN 39
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,143,903
Number of Sequences: 63676
Number of extensions: 272342
Number of successful extensions: 1672
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 1627
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1636
length of query: 468
length of database: 12,639,632
effective HSP length: 101
effective length of query: 367
effective length of database: 6,208,356
effective search space: 2278466652
effective search space used: 2278466652
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144389.8


