

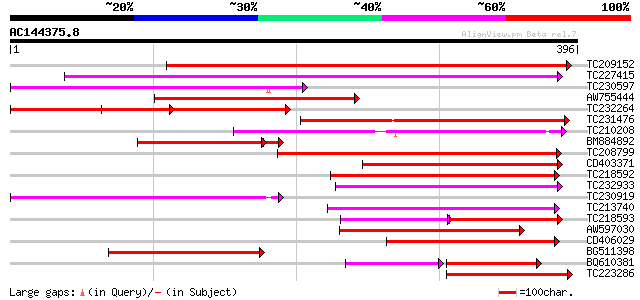
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144375.8 + phase: 0
(396 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209152 weakly similar to PIR|B96764|B96764 protein integral me... 447 e-126
TC227415 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (89%) 243 1e-64
TC230597 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g1... 207 9e-54
AW755444 189 2e-48
TC232264 weakly similar to UP|Q941E1 (Q941E1) At1g66760/F4N21_11... 187 5e-48
TC231476 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g1... 185 3e-47
TC210208 169 2e-42
BM884892 153 2e-39
TC208799 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, pa... 152 3e-37
CD403371 144 5e-35
TC218592 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, pa... 141 4e-34
TC232933 similar to GB|AAN73299.1|25141209|BT002302 At3g26590/MF... 128 4e-30
TC230919 110 1e-24
TC213740 weakly similar to UP|TT12_ARATH (Q9LYT3) TRANSPARENT TE... 109 2e-24
TC218593 81 9e-24
AW597030 similar to SP|Q9LYT3|TT12 TRANSPARENT TESTA 12 protein.... 102 4e-22
CD406029 101 7e-22
BG511398 95 6e-20
BQ610381 66 1e-19
TC223286 similar to GB|AAP31960.1|30387589|BT006616 At1g15170 {A... 91 9e-19
>TC209152 weakly similar to PIR|B96764|B96764 protein integral membrane
protein F25P22.12 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (41%)
Length = 1166
Score = 447 bits (1149), Expect = e-126
Identities = 213/283 (75%), Positives = 247/283 (87%)
Frame = +1
Query: 110 ICWCLVFKFGLGHVGAAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLLYIAEFC 169
ICW LVFK GLGHVGAA AIG++YWLNV+WL IYM YSPAC+KTKIVFS N+LL I EF
Sbjct: 1 ICWGLVFKLGLGHVGAALAIGVSYWLNVVWLAIYMIYSPACQKTKIVFSSNALLSIPEFL 180
Query: 170 QFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLETSVLSVCLSTTTLHYFIPHAIGASAST 229
+ AIPSGLMFC EWWSFE+LT++AG+LPN QLET+VLS+CL+TTTLHYFIP+A+GASAST
Sbjct: 181 KLAIPSGLMFCFEWWSFEVLTLLAGILPNPQLETAVLSICLNTTTLHYFIPYAVGASAST 360
Query: 230 RVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDM 289
RVSNELGAGNP+ AKGAVRV VI+G+AEA IVST+F+ R+++G AYSNDKEV+DYV +M
Sbjct: 361 RVSNELGAGNPKTAKGAVRVVVILGVAEAAIVSTVFISCRHVLGYAYSNDKEVIDYVAEM 540
Query: 290 VPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGL 349
P LCVSV+ADS+I ALSGIARGGGFQ IGAYVNLGAYYLVG P+ LGF L+L AKGL
Sbjct: 541 APLLCVSVTADSLIGALSGIARGGGFQEIGAYVNLGAYYLVGIPMGLLLGFHLQLRAKGL 720
Query: 350 WMGTLTGSILNVIILAVVTMLTDWQKEATKARERIAEKPIEAH 392
WMGTL+GS+ VIILA+VT L DWQKEATKARER+ E I+AH
Sbjct: 721 WMGTLSGSLTQVIILAIVTALIDWQKEATKARERVVENSIKAH 849
>TC227415 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (89%)
Length = 1748
Score = 243 bits (620), Expect = 1e-64
Identities = 124/348 (35%), Positives = 206/348 (58%)
Frame = +2
Query: 39 ISLMWIFIDKLLLLFGQDIEIAQAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVF 98
++++++F + +L+ G+ IA AA + LIP +F +A + ++ Q QS++ P +
Sbjct: 548 LTVIYVFSEPMLIFLGESPRIASAAALFVYGLIPQIFAYAANFPIQKFLQAQSIVAPSAY 727
Query: 99 SSIVILCLHVPICWCLVFKFGLGHVGAAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFS 158
S L +H+ + W V++ GLG +GA+ + +++W+ VI +Y+ S C +T F+
Sbjct: 728 ISAATLVVHLGMSWVAVYEIGLGLLGASLVLSLSWWIMVIGQYVYIVKSERCRRTWQGFT 907
Query: 159 YNSLLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLETSVLSVCLSTTTLHYF 218
+ + + F + + S +M CLE W F+IL ++AGLLPN +L LS+C + + +
Sbjct: 908 WEAFSGLYGFFKLSAASAVMLCLETWYFQILVLLAGLLPNPELALDSLSICTTISGWVFM 1087
Query: 219 IPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSN 278
I A+AS RVSNELGA +P++A +V V +I +VI + + L R++I A++
Sbjct: 1088ISVGFNAAASVRVSNELGARSPKSASFSVVVVTVISFIISVIAALVVLALRDVISYAFTG 1267
Query: 279 DKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFL 338
+EV V+D+ P L +S+ + I LSG+A G G+Q AYVN+G YY VG P+ L
Sbjct: 1268GEEVAAAVSDLCPLLALSLVLNGIQPVLSGVAVGCGWQAFVAYVNVGCYYGVGIPLGAVL 1447
Query: 339 GFGLKLNAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERIAE 386
GF + AKG+W+G L G+++ IIL VT TDW KE +A +R+ +
Sbjct: 1448GFYFQFGAKGIWLGMLGGTVMQTIILLWVTFRTDWTKEVEEAAKRLTK 1591
Score = 36.2 bits (82), Expect = 0.027
Identities = 20/65 (30%), Positives = 34/65 (51%)
Frame = +1
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
M A+ETLCGQ +GA+++ +G Y+ + I L SL + +D+ + + D I
Sbjct: 433 MGSAVETLCGQAFGAQKYGMLGVYMQRSTILL-------SLAGVVVDRDIRIQRTDANIP 591
Query: 61 QAARE 65
+ E
Sbjct: 592 RGITE 606
>TC230597 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g15170
{Arabidopsis thaliana;} , partial (51%)
Length = 1061
Score = 207 bits (526), Expect = 9e-54
Identities = 110/259 (42%), Positives = 151/259 (57%), Gaps = 51/259 (19%)
Frame = +3
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
MA LET+CGQ YG +++ +IG +A+ +LILV P+SL+WI ++ +L+ GQD I+
Sbjct: 249 MASGLETICGQAYGGQQYQRIGIQTYTAIFSLILVSIPVSLLWINMETILVFIGQDPLIS 428
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGL 120
A ++ I L+PALF +A+LQ L+RYFQ+QS++ PM SS V L +HVP+CW LVFK L
Sbjct: 429 HEAGKFTIWLVPALFAYAILQPLVRYFQVQSLLLPMFASSCVTLIIHVPLCWALVFKTRL 608
Query: 121 GHVGAAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLM-- 178
+VG A A+ I+ W NVI+LG+YM+Y AC KT+ S + EF +FAIPS +M
Sbjct: 609 SNVGGALAVSISIWSNVIFLGLYMRYFSACAKTRAPISMELFKGMWEFFRFAIPSAVMVW 788
Query: 179 ------------------------------------------FC-------LEWWSFEIL 189
FC LEWWS+E+L
Sbjct: 789 *LFFLFNQYFTYSCYQITKSFLVQQIFYNVLYCFSKTSI*HNFCIHVDDGSLEWWSYELL 968
Query: 190 TIVAGLLPNSQLETSVLSV 208
+++GLLPN QLETSVLSV
Sbjct: 969 VLLSGLLPNPQLETSVLSV 1025
>AW755444
Length = 432
Score = 189 bits (480), Expect = 2e-48
Identities = 90/143 (62%), Positives = 109/143 (75%)
Frame = +1
Query: 102 VILCLHVPICWCLVFKFGLGHVGAAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFSYNS 161
V+L LH+PICW LVF GLG GAA +IGI+YWL+V+ L IY KY P+C+KTKI N+
Sbjct: 1 VVLVLHIPICWVLVFGLGLGQNGAAISIGISYWLSVMLLLIYTKYYPSCQKTKIALGSNA 180
Query: 162 LLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLETSVLSVCLSTTTLHYFIPH 221
L I EF AIPS LM C EWWSFE++ I+AGLLPN +LETSVLS+CL+ TLHYFIP+
Sbjct: 181 LRSIKEFFFLAIPSALMICFEWWSFELVVILAGLLPNPKLETSVLSICLNICTLHYFIPY 360
Query: 222 AIGASASTRVSNELGAGNPRAAK 244
GA+ STRVSNEL A P+AA+
Sbjct: 361 GTGAAVSTRVSNELXARRPQAAR 429
>TC232264 weakly similar to UP|Q941E1 (Q941E1) At1g66760/F4N21_11, partial
(58%)
Length = 701
Score = 187 bits (476), Expect = 5e-48
Identities = 90/115 (78%), Positives = 98/115 (84%)
Frame = +1
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
M+GALETLCGQTYGAEE+ K GNY A++TL LVC PISL+WIF DK+LLLF QD EI+
Sbjct: 166 MSGALETLCGQTYGAEEYRKFGNYTWCAIVTLTLVCLPISLVWIFTDKILLLFSQDPEIS 345
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLV 115
AAREYCI LIPALFGHAVLQ+L RYFQ QSMIFPMVFSSI LCLHVPICW LV
Sbjct: 346 HAAREYCIYLIPALFGHAVLQALTRYFQTQSMIFPMVFSSITALCLHVPICWSLV 510
Score = 118 bits (296), Expect = 4e-27
Identities = 60/133 (45%), Positives = 83/133 (62%), Gaps = 1/133 (0%)
Frame = +2
Query: 65 EYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGLGHVG 124
+YC CL+ L ++Q + + + M+F + + + + F H
Sbjct: 308 KYCCCLVKTL--RFLMQLVSTAYTSFQLYLVMLFFKLSLATSRLRV*SFPWFSAQSPHCV 481
Query: 125 AAFA-IGIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLMFCLEW 183
F +G++YWLNV+WL IYM YSPAC+KTKIVFS N+LL I EF + AIPSGLMFC EW
Sbjct: 482 CMFLFVGVSYWLNVVWLAIYMIYSPACQKTKIVFSSNALLSIPEFLKLAIPSGLMFCFEW 661
Query: 184 WSFEILTIVAGLL 196
WSFE+LT++AG+L
Sbjct: 662 WSFEVLTLLAGIL 700
>TC231476 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g15170
{Arabidopsis thaliana;} , partial (32%)
Length = 734
Score = 185 bits (470), Expect = 3e-47
Identities = 97/189 (51%), Positives = 129/189 (67%), Gaps = 1/189 (0%)
Frame = +2
Query: 204 SVLSVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVS- 262
SVLS+CL+T + + IP I A+ASTR+SNELGAGNP AA AV A+ I E IVS
Sbjct: 2 SVLSICLNTISTLFSIPFGIAAAASTRISNELGAGNPHAAHVAVLAAMSFAIMETAIVSG 181
Query: 263 TLFLCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYV 322
TLF+C R+ G +SN+KEVVDYVT M P +C+SV DSI L+G+ARG G+Q IG YV
Sbjct: 182 TLFVC-RHDFGYIFSNEKEVVDYVTVMAPLICISVILDSIQGVLAGVARGCGWQHIGVYV 358
Query: 323 NLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARE 382
NLGA+YL G P+A L F K+ KGLW+G G+ + I+ + +T +W+++A KAR+
Sbjct: 359 NLGAFYLCGIPVAATLAFLAKMRGKGLWIGVQVGAFVQCILFSTITSCINWEQQAIKARK 538
Query: 383 RIAEKPIEA 391
R+ + I A
Sbjct: 539 RLFDSEISA 565
>TC210208
Length = 743
Score = 169 bits (428), Expect = 2e-42
Identities = 91/240 (37%), Positives = 139/240 (57%), Gaps = 7/240 (2%)
Frame = +2
Query: 157 FSYNSLLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLETSVLSVCLSTTTLH 216
FS +S Y+ + A+PS M CLE+W+FE+L +AGL+P+SQ+ TS++++C++T +
Sbjct: 44 FSTHSFRYVFTNMRLALPSAAMVCLEYWAFEVLVFLAGLMPDSQITTSLIAICINTEFIA 223
Query: 217 YFIPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCF-------R 269
Y I + + A+ASTRVSNELGAGNP AK A+ V + + + L LCF
Sbjct: 224 YMITYGLSAAASTRVSNELGAGNPERAKHAMSVTLKLSL-------LLGLCFVLALGFGH 382
Query: 270 NIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYL 329
NI +S+ + + P L +S+ D+I LSG++RG G+Q + AY+NL +YL
Sbjct: 383 NIWIQFFSDSSTIKKEFASVTPLLAISILLDAIQGVLSGVSRGCGWQHLAAYINLATFYL 562
Query: 330 VGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERIAEKPI 389
+G PI+ FLGF L KGLW+G + G + L + W K +R+ E+P+
Sbjct: 563 IGLPISCFLGFKTNLQYKGLWIGLICGLLCQSGTLFLFIRRKXWTK-LDLSRDNDKERPL 739
>BM884892
Length = 421
Score = 153 bits (387), Expect(2) = 2e-39
Identities = 72/90 (80%), Positives = 78/90 (86%)
Frame = +1
Query: 90 QSMIFPMVFSSIVILCLHVPICWCLVFKFGLGHVGAAFAIGIAYWLNVIWLGIYMKYSPA 149
QSMIFPMVFSSI LCLHVPICW LVFK GLGHVGAA AIG++YWLNV+WL IYM YSPA
Sbjct: 1 QSMIFPMVFSSITALCLHVPICWGLVFKLGLGHVGAALAIGVSYWLNVVWLAIYMIYSPA 180
Query: 150 CEKTKIVFSYNSLLYIAEFCQFAIPSGLMF 179
C+KTKIVFS N+LL I EF + AIPSGLMF
Sbjct: 181 CQKTKIVFSSNALLSIPEFLKLAIPSGLMF 270
Score = 27.3 bits (59), Expect(2) = 2e-39
Identities = 8/14 (57%), Positives = 12/14 (85%)
Frame = +2
Query: 178 MFCLEWWSFEILTI 191
++ EWWSFE+LT+
Sbjct: 380 VYSFEWWSFEVLTL 421
>TC208799 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, partial (46%)
Length = 924
Score = 152 bits (384), Expect = 3e-37
Identities = 77/198 (38%), Positives = 124/198 (61%)
Frame = +2
Query: 188 ILTIVAGLLPNSQLETSVLSVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRAAKGAV 247
IL ++ G + N+++E LS+CL+ I A+AS RV+NELG G+ +AAK ++
Sbjct: 11 ILVLLTGNMKNAEVEIDALSICLNINGWEMMISLGFMAAASVRVANELGRGSAKAAKFSI 190
Query: 248 RVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICALS 307
V+V+ +A ++ FL FR + ++++KEV V D+ P L VS+ +S+ LS
Sbjct: 191 IVSVLTSLAIGFLLFIFFLFFRERLAYIFTSNKEVAFAVGDLSPLLSVSILLNSVQPVLS 370
Query: 308 GIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAVV 367
G+A G G+Q+I AYVN+G YY +G P+ LG L L KG+W+G L G+++ I+L V+
Sbjct: 371 GVAIGAGWQSIVAYVNMGCYYAIGIPVGIVLGNVLDLQVKGIWIGMLFGTLIQTIVLIVI 550
Query: 368 TMLTDWQKEATKARERIA 385
T T+W ++ T A++RI+
Sbjct: 551 TYKTNWDEQVTIAQKRIS 604
>CD403371
Length = 592
Score = 144 bits (364), Expect = 5e-35
Identities = 67/140 (47%), Positives = 94/140 (66%)
Frame = -1
Query: 247 VRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICAL 306
V +++ +AV+ S++ CFR+++G A+SN+ EVV YV +VP LC+S D + L
Sbjct: 592 VFAVIVLAFTDAVVFSSVLFCFRHVLGFAFSNEMEVVHYVAKIVPVLCLSFMVDGFLGVL 413
Query: 307 SGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAV 366
GI RG G+Q IGA NL AYY VG P++ FGL N KGLW+G LTGS L IILA+
Sbjct: 412 CGIVRGSGWQKIGAITNLVAYYAVGIPVSLLFXFGLNFNGKGLWIGILTGSTLQTIILAL 233
Query: 367 VTMLTDWQKEATKARERIAE 386
+T T+W+K+A+ A ER++E
Sbjct: 232 LTAFTNWEKQASLAIERLSE 173
>TC218592 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, partial (37%)
Length = 715
Score = 141 bits (356), Expect = 4e-34
Identities = 67/160 (41%), Positives = 104/160 (64%)
Frame = +3
Query: 225 ASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVD 284
A+AS RV+NELG G+ +AAK ++ V V+ +A ++ FL R + ++++K+V D
Sbjct: 18 AAASVRVANELGKGSSKAAKFSIVVTVLTSLAIGFVLFLFFLFLRGKLAYIFTSNKDVAD 197
Query: 285 YVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKL 344
V D+ P L +S+ +S+ LSG+A G G+Q+I AYVN+G YY++G P+ LG L L
Sbjct: 198 AVGDLSPLLAISILLNSVQPVLSGVAIGAGWQSIVAYVNIGCYYIIGIPVGVVLGNVLNL 377
Query: 345 NAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERI 384
KG+W+G L G+ + ++L V+T TDW ++ TKAR RI
Sbjct: 378 QVKGIWIGMLFGTFIQTVVLTVITYKTDWDEQVTKARNRI 497
>TC232933 similar to GB|AAN73299.1|25141209|BT002302 At3g26590/MFE16_11
{Arabidopsis thaliana;} , partial (30%)
Length = 545
Score = 128 bits (322), Expect = 4e-30
Identities = 65/159 (40%), Positives = 96/159 (59%)
Frame = +1
Query: 228 STRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVT 287
S RVSNELGA +PR AK ++ VA+I V++S + + FRN +SND EV V
Sbjct: 55 SVRVSNELGASHPRTAKFSLLVAMITSTLIGVMLSMVLIIFRNHYPFFFSNDSEVRAMVV 234
Query: 288 DMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAK 347
++ P L + + +++ LSG+A G G+Q + AYVN+ YYL G P+ LG+ L +
Sbjct: 235 ELTPMLALCIVINNVQPVLSGVAVGAGWQALVAYVNIACYYLFGIPLGLVLGYKLDMGVM 414
Query: 348 GLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERIAE 386
G+W G L+G+IL +L + TDW KEA+ A +RI +
Sbjct: 415 GIWSGMLSGTILQTCVLFFLVYRTDWNKEASLAEDRIKQ 531
>TC230919
Length = 1034
Score = 110 bits (275), Expect = 1e-24
Identities = 62/191 (32%), Positives = 108/191 (56%)
Frame = +2
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
++GALETL Q +GAEE+ +G Y+ ++ I ++ IS++W + + +L+L Q +IA
Sbjct: 29 LSGALETLXXQGFGAEEYQMLGIYLQASCIISLIFSIIISIIWFYTEPILVLLHQSQDIA 208
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGL 120
+ Y LIP LF + LQ+++R+ Q QS++ +V S + L +H+ I L+F L
Sbjct: 209 RTTSLYTKFLIPGLFALSFLQNILRFLQTQSVVKSLVVFSAIPLLVHIFIA*ALIFCTDL 388
Query: 121 GHVGAAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLMFC 180
+GA A+ I+ W+++ L +Y+ Y+ +T FS+ S YI + A+ S M
Sbjct: 389 SFIGAPVAVSISLWISIPLLVMYIMYAERFRQTWTGFSFESFNYIFTDLKLALLSAAMV- 565
Query: 181 LEWWSFEILTI 191
W+ + LT+
Sbjct: 566 --WYVSKNLTL 592
>TC213740 weakly similar to UP|TT12_ARATH (Q9LYT3) TRANSPARENT TESTA 12
protein, partial (20%)
Length = 733
Score = 109 bits (272), Expect = 2e-24
Identities = 55/162 (33%), Positives = 89/162 (53%)
Frame = +3
Query: 223 IGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEV 282
I + S RVSN LG +PRAA + V + + ++ T+ ++ +++ +++
Sbjct: 39 INTAISVRVSNTLGMSHPRAAIYSFCVTMFQSLLLGILFMTVIFFSKDEFAKIFTDSEDM 218
Query: 283 VDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGL 342
+ D+ L V++ +S +SG+A G G+Q + Y+NL YY+VG PI FLGF L
Sbjct: 219 ILAAADLAYLLGVTIVLNSASQVMSGVAIGSGWQVMVGYINLACYYIVGLPIGIFLGFKL 398
Query: 343 KLNAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERI 384
L KGLW GT+ GSIL ++L + T+W KE + R+
Sbjct: 399 HLGVKGLWGGTMCGSILQTLVLFTIIWKTNWSKEVEQTAHRM 524
>TC218593
Length = 991
Score = 81.3 bits (199), Expect(2) = 9e-24
Identities = 34/79 (43%), Positives = 54/79 (68%)
Frame = +2
Query: 308 GIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAVV 367
G+A G G+Q+I AYVN+G YYL+G P+ LG + L KG+W+G L G+++ I+L ++
Sbjct: 341 GVAVGAGWQSIVAYVNIGCYYLIGIPVGIVLGNIIHLQVKGIWIGMLFGTLIQTIVLTII 520
Query: 368 TMLTDWQKEATKARERIAE 386
T T+W ++ AR RI++
Sbjct: 521 TYKTNWDEQVIIARNRISK 577
Score = 47.0 bits (110), Expect(2) = 9e-24
Identities = 26/78 (33%), Positives = 45/78 (57%)
Frame = +3
Query: 232 SNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVP 291
+NELG G+ + AK ++ V V+ + I+ LFL R + ++++++V V D+ P
Sbjct: 3 ANELGRGSSKDAKFSIVVTVLTSFSIGFILFVLFLFLREKVAYLFTSNEDVATAVGDLSP 182
Query: 292 FLCVSVSADSIICALSGI 309
L VS+ +SI LSG+
Sbjct: 183 LLAVSLLLNSIQPVLSGM 236
>AW597030 similar to SP|Q9LYT3|TT12 TRANSPARENT TESTA 12 protein. [Mouse-ear
cress] {Arabidopsis thaliana}, partial (25%)
Length = 401
Score = 102 bits (253), Expect = 4e-22
Identities = 52/129 (40%), Positives = 78/129 (60%)
Frame = +1
Query: 231 VSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMV 290
VSNELGA +PR AK +V V I +V+ T+ L FR + +++D +V+D V+++
Sbjct: 10 VSNELGASHPRVAKFSVFVVNGTSILISVVFCTIILIFRVSLSKLFTSDSDVIDAVSNLT 189
Query: 291 PFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLW 350
P L +SV + I LSG+A G G+Q + AYVNL +YY+VG + LGF L G+W
Sbjct: 190 PLLAISVFFNGIQPILSGVAIGSGWQALVAYVNLASYYVVGLTVGCVLGFKTSLGVAGIW 369
Query: 351 MGTLTGSIL 359
G + G ++
Sbjct: 370 WGMILGVLI 396
>CD406029
Length = 464
Score = 101 bits (251), Expect = 7e-22
Identities = 49/121 (40%), Positives = 77/121 (63%)
Frame = -2
Query: 264 LFLCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVN 323
LFL R + +++++VV V D+ P L +S+ +SI LSG+A G G+Q+ AYVN
Sbjct: 457 LFLILREKVAYLLTSNEDVVTAVGDLSPLLALSLLLNSIQPVLSGVALGPGWQSTVAYVN 278
Query: 324 LGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARER 383
+G YYL+G P+ LG + L KG+W+G L G+++ IIL ++T T+W ++ AR+R
Sbjct: 277 IGCYYLIGIPVGIVLGNIIHLQVKGIWIGMLFGTLIQTIILIIITYKTNWDEQVIIARDR 98
Query: 384 I 384
I
Sbjct: 97 I 95
>BG511398
Length = 425
Score = 94.7 bits (234), Expect = 6e-20
Identities = 43/109 (39%), Positives = 69/109 (62%)
Frame = -1
Query: 70 LIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGLGHVGAAFAI 129
+IP+LF + +L+ ++++ Q Q ++FPMV +S + LHV CW LVFK GL + GAA A
Sbjct: 398 MIPSLFAYGILRCILKFLQTQKIVFPMVLTSGIAAVLHVLFCWLLVFKSGLANRGAALAN 219
Query: 130 GIAYWLNVIWLGIYMKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLM 178
I+YW+N I + +Y++ S AC+ + FS +L + +F + PS M
Sbjct: 218 SISYWVNAILISLYVRLSSACKHSWTGFSKMALHNLLDFLKLGAPSAAM 72
>BQ610381
Length = 411
Score = 66.2 bits (160), Expect(2) = 1e-19
Identities = 29/66 (43%), Positives = 42/66 (62%)
Frame = +1
Query: 306 LSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILA 365
LSG+A G G+Q + AYVN+ YYL G P+ LG+ L KG+W+G ++G+IL +L
Sbjct: 214 LSGVAIGAGWQALVAYVNIACYYLFGIPVGLVLGYKLDWGVKGIWLGMISGTILQTCVLL 393
Query: 366 VVTMLT 371
V+ T
Sbjct: 394 VLIYKT 411
Score = 48.1 bits (113), Expect(2) = 1e-19
Identities = 24/69 (34%), Positives = 41/69 (58%)
Frame = +2
Query: 235 LGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVTDMVPFLC 294
L A +PR A ++ VAVI + V+++ + + RN + +SND EV D V D+ PFLC
Sbjct: 2 LRARHPRTALFSLVVAVITSVLIGVLLAIVLMISRNEYPSLFSNDTEVQDLVKDLTPFLC 181
Query: 295 VSVSADSII 303
+ ++++
Sbjct: 182 FCIVINNVL 208
>TC223286 similar to GB|AAP31960.1|30387589|BT006616 At1g15170 {Arabidopsis
thaliana;} , partial (17%)
Length = 414
Score = 90.9 bits (224), Expect = 9e-19
Identities = 42/88 (47%), Positives = 61/88 (68%)
Frame = +3
Query: 306 LSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILA 365
L+GIARG G+Q IG YVNLGA+YL G P+A L F K++ KGLW+G G+ + +L+
Sbjct: 9 LAGIARGCGWQHIGVYVNLGAFYLCGIPVAASLAFLAKMSGKGLWIGLQVGAFVQCALLS 188
Query: 366 VVTMLTDWQKEATKARERIAEKPIEAHD 393
VT T+W+++A KAR+R+ + I A +
Sbjct: 189 TVTSCTNWEQQAMKARKRLFDSEISAEN 272
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.329 0.142 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,055,281
Number of Sequences: 63676
Number of extensions: 330650
Number of successful extensions: 2709
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 2673
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2704
length of query: 396
length of database: 12,639,632
effective HSP length: 99
effective length of query: 297
effective length of database: 6,335,708
effective search space: 1881705276
effective search space used: 1881705276
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144375.8


