

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144375.4 - phase: 0
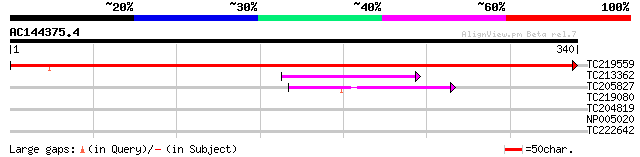
(340 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219559 similar to UP|Q9SXN4 (Q9SXN4) SLL2-S9-protein, partial ... 589 e-169
TC213362 similar to UP|Q6ID86 (Q6ID86) At4g34360, partial (59%) 53 2e-07
TC205827 42 3e-04
TC219080 similar to UP|Q9LUT4 (Q9LUT4) Gb|AAF34859.1, partial (45%) 33 0.14
TC204819 UP|Q43445 (Q43445) S-adenosyl-L-methionine:delta24-ster... 28 4.6
NP005020 S-adenosyl-L-methionine:delta24-sterol-C-methyltransferase 28 4.6
TC222642 similar to GB|AAP42724.1|30984522|BT008711 At5g17660 {A... 28 7.8
>TC219559 similar to UP|Q9SXN4 (Q9SXN4) SLL2-S9-protein, partial (86%)
Length = 1124
Score = 589 bits (1519), Expect = e-169
Identities = 293/342 (85%), Positives = 316/342 (91%), Gaps = 2/342 (0%)
Frame = +3
Query: 1 MAGIRLQPEDSDVSQQARAVAL--VDLVSDDDRSIAADSWSIKSEYGSTLDDDQRHADAA 58
MAGIRLQPEDSDVSQQ+RAVAL DLVSDDDRS+AADSWSIKSEYGSTLDDDQRHADAA
Sbjct: 33 MAGIRLQPEDSDVSQQSRAVALSATDLVSDDDRSVAADSWSIKSEYGSTLDDDQRHADAA 212
Query: 59 EALSNVNLRAASDYSSDKDEPDAEAVSSMLGFQSYWDAAYTDELTNFHEHGHAGEVWFGD 118
EALSN NLR SDYSSDKDEPD+EAV+SMLGFQSYWD+AY DELTNF EHGH GEVWFG
Sbjct: 213 EALSNANLRPPSDYSSDKDEPDSEAVTSMLGFQSYWDSAYADELTNFREHGHTGEVWFGV 392
Query: 119 NVMEVVASWTKTLCIDISQGRLPNHVDDVKADAGELDDKLLSSWNVLDIGTGNGLLLQEL 178
+VMEVVASWTK LC++ISQG +PN VD+VKA+A +L DK+LS+W+VLDIGTGNGLLLQEL
Sbjct: 393 DVMEVVASWTKALCVEISQGHIPNGVDEVKAEADKLGDKVLSTWSVLDIGTGNGLLLQEL 572
Query: 179 AKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETKLEQVFQLVMDKGTLDA 238
AKQGFSDLTGTDYSERAI+LAQSLANRDGF N+KFLVDDVLETKLEQ F+LVMDKGTLDA
Sbjct: 573 AKQGFSDLTGTDYSERAISLAQSLANRDGFSNVKFLVDDVLETKLEQEFRLVMDKGTLDA 752
Query: 239 IGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNSTKDELVQEVESFNQRKSATAPEPEV 298
IGLHPDGPVKRMMYWDSVSRLVA GGILV+TSCN+TKDELVQEVESFNQRK ATA E
Sbjct: 753 IGLHPDGPVKRMMYWDSVSRLVASGGILVITSCNNTKDELVQEVESFNQRKIATAQELGA 932
Query: 299 AKDDESCRDPLFQYVSHVRTYPTFMFGGSVGSRVATVAFLRK 340
KD+E CR+P FQYV+HVRTYPTFMFGGSVGSRVATVAFLRK
Sbjct: 933 VKDEEPCREPPFQYVNHVRTYPTFMFGGSVGSRVATVAFLRK 1058
>TC213362 similar to UP|Q6ID86 (Q6ID86) At4g34360, partial (59%)
Length = 491
Score = 52.8 bits (125), Expect = 2e-07
Identities = 30/84 (35%), Positives = 47/84 (55%), Gaps = 1/84 (1%)
Frame = +1
Query: 164 VLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETKL 223
VL++G GN + ++L K G +++T D S A+ Q GF +IK L D+LE
Sbjct: 178 VLELGCGNSQMCEQLHKDGTTNITCIDLSPGAVQNMQKRLLSRGFKDIKVLQADMLELPF 357
Query: 224 E-QVFQLVMDKGTLDAIGLHPDGP 246
E + F LV++KGT+D + + P
Sbjct: 358 EDECFDLVIEKGTMDVLFVDSGDP 429
>TC205827
Length = 2695
Score = 42.4 bits (98), Expect = 3e-04
Identities = 33/103 (32%), Positives = 52/103 (50%), Gaps = 3/103 (2%)
Frame = +1
Query: 168 GTGNGLLLQELAKQGFSDLTGTDYSERAIN--LAQSLANRDGFPNIKFLVDDVLETKLE- 224
G GN L + L G + +T D+S+ I L +++ +R P +++ V D+ + E
Sbjct: 226 GCGNSRLSEHLYDAGHTAITNIDFSKVVIGDMLRRNVRDR---PLMRWRVMDMTVMQFED 396
Query: 225 QVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILV 267
+ F V+DKG LDA+ GP Y V R++ PGG V
Sbjct: 397 ESFGAVIDKGGLDALMEPELGPKLGNQYLSEVKRVLKPGGKFV 525
>TC219080 similar to UP|Q9LUT4 (Q9LUT4) Gb|AAF34859.1, partial (45%)
Length = 1105
Score = 33.5 bits (75), Expect = 0.14
Identities = 28/110 (25%), Positives = 49/110 (44%), Gaps = 3/110 (2%)
Frame = +1
Query: 164 VLDIGTGNGLLLQELAKQG-FSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETK 222
VL +G GN + + G ++D+ D S I ++ P +KF+ DV +
Sbjct: 247 VLVVGCGNSAFSEGMVVDGGYTDVVNIDISSVVIEAMKT--KHQDCPQLKFMKMDVRDMS 420
Query: 223 LEQV--FQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTS 270
Q F V+DKGTLD+I + + + R++ G+ V+ +
Sbjct: 421 DFQSGSFGAVIDKGTLDSILCGNNSRQNATKMLEEIWRVLKDKGVYVLVT 570
>TC204819 UP|Q43445 (Q43445)
S-adenosyl-L-methionine:delta24-sterol-C-
methyltransferase , partial (98%)
Length = 1337
Score = 28.5 bits (62), Expect = 4.6
Identities = 25/110 (22%), Positives = 44/110 (39%), Gaps = 5/110 (4%)
Frame = +1
Query: 98 YTDELTNFHEHG-----HAGEVWFGDNVMEVVASWTKTLCIDISQGRLPNHVDDVKADAG 152
Y D +T+F+E G H W G+++ E + L + +
Sbjct: 226 YYDLVTSFYEFGWGESFHFAPRWKGESLRESIKRHEHFLPLQLG---------------- 357
Query: 153 ELDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSL 202
L VLD+G G G L+E+++ + +TG + +E I + L
Sbjct: 358 -----LKPGQKVLDVGCGIGGPLREISRFSSTSITGLNNNEYQITRGKEL 492
>NP005020 S-adenosyl-L-methionine:delta24-sterol-C-methyltransferase
Length = 1104
Score = 28.5 bits (62), Expect = 4.6
Identities = 25/110 (22%), Positives = 44/110 (39%), Gaps = 5/110 (4%)
Frame = +1
Query: 98 YTDELTNFHEHG-----HAGEVWFGDNVMEVVASWTKTLCIDISQGRLPNHVDDVKADAG 152
Y D +T+F+E G H W G+++ E + L + +
Sbjct: 223 YYDLVTSFYEFGWGESFHFAPRWKGESLRESIKRHEHFLPLQLG---------------- 354
Query: 153 ELDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSL 202
L VLD+G G G L+E+++ + +TG + +E I + L
Sbjct: 355 -----LKPGQKVLDVGCGIGGPLREISRFSSTSITGLNNNEYQITRGKEL 489
>TC222642 similar to GB|AAP42724.1|30984522|BT008711 At5g17660 {Arabidopsis
thaliana;} , partial (64%)
Length = 1094
Score = 27.7 bits (60), Expect = 7.8
Identities = 31/142 (21%), Positives = 54/142 (37%), Gaps = 12/142 (8%)
Frame = +1
Query: 85 SSMLGFQSYWDAAYTDELTNFHEHGHAGEVWFGDNVMEVVASWTKTLCIDISQGRLPNHV 144
SS L F S + + +LT + V A + +D+ R+ HV
Sbjct: 61 SSSLSFSSNCHVSSSAQLTTLQQQQQQPLPRSSQLVALEYADLNLSHNLDLGHVRIRQHV 240
Query: 145 DDVKADAGELDDKLLSSWN----------VLDIGTGNGLLLQELAKQGFSD--LTGTDYS 192
+ + + + WN ++DIG G+G L LAK+ + G +
Sbjct: 241 NPLSSSLSV--PAQVPEWNHAFADPTLPLMVDIGCGSGRFLMWLAKRTPKERNFLGLEIR 414
Query: 193 ERAINLAQSLANRDGFPNIKFL 214
++ + A+S A N+ FL
Sbjct: 415 QKIVKRAESWAKDLALDNVHFL 480
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,275,160
Number of Sequences: 63676
Number of extensions: 164185
Number of successful extensions: 503
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 501
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 502
length of query: 340
length of database: 12,639,632
effective HSP length: 98
effective length of query: 242
effective length of database: 6,399,384
effective search space: 1548650928
effective search space used: 1548650928
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144375.4


