

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144345.3 + phase: 0
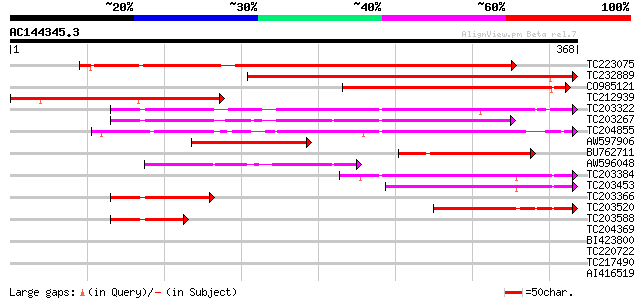
(368 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC223075 UP|MEP1_SOYBN (P29136) Metalloendoproteinase 1 precurso... 306 7e-84
TC232889 weakly similar to UP|O65340 (O65340) Metalloproteinase ... 293 8e-80
CO985121 219 2e-57
TC212939 weakly similar to UP|Q9ZUJ5 (Q9ZUJ5) T2K10.2 protein, p... 189 1e-48
TC203322 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 140 9e-34
TC203267 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 132 3e-31
TC204855 UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2, complete 127 8e-30
AW597906 weakly similar to PIR|T00643|T006 zinc metalloproteinas... 126 2e-29
BU762711 105 2e-23
AW596048 weakly similar to GP|16901508|gb| matrix metalloprotein... 86 2e-17
TC203384 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 71 9e-13
TC203453 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 66 2e-11
TC203366 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 64 8e-11
TC203520 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 59 3e-09
TC203588 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 46 3e-05
TC204369 similar to GB|BAB09857.1|9759402|AB008268 phi-1-like pr... 33 0.16
BI423800 similar to GP|27902363|gb| polyphenol oxidase {Trifoliu... 30 1.7
TC220722 similar to UP|Q9LM55 (Q9LM55) F2E2.11, partial (28%) 30 1.7
TC217490 similar to UP|Q9FNN8 (Q9FNN8) Cleft lip and palate asso... 30 1.7
AI416519 30 2.3
>TC223075 UP|MEP1_SOYBN (P29136) Metalloendoproteinase 1 precursor (SMEP1) ,
complete
Length = 1198
Score = 306 bits (785), Expect = 7e-84
Identities = 157/287 (54%), Positives = 193/287 (66%), Gaps = 3/287 (1%)
Frame = +3
Query: 46 PGAWDG---YKNFTGCSRGKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRT 102
P AWDG YK FT G+ Y GLS +KNYF+H GYIPN P +F D+FD+ L SA++T
Sbjct: 135 PYAWDGEATYK-FTTYHPGQNYKGLSNVKNYFHHLGYIPNAP--HFDDNFDDTLVSAIKT 305
Query: 103 YQKNFNLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFP 162
YQKN+NLN+TG+ D T+ I+ PRCGV DII T + +T+F ++ Y+FF
Sbjct: 306 YQKNYNLNVTGKFDINTLKQIMTPRCGVPDIIINT---------NKTTSFGMISDYTFFK 458
Query: 163 GQPRWPEGTQTLTYAFDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGF 222
PRW GT LTYAF P LDD K A AF++W+ V I F E TSY +++IKI F
Sbjct: 459 DMPRWQAGTTQLTYAFSPEPRLDDTFKSAIARAFSKWTPVVNIAFQETTSYETANIKILF 638
Query: 223 YSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVH 282
S +HGD FDG G L HAF+PTDGR H D E WV +GDVT+S +++A DLESV VH
Sbjct: 639 ASKNHGDPYPFDGPGGILGHAFAPTDGRCHFDADEYWVASGDVTKSPVTSAFDLESVAVH 818
Query: 283 EIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNP 329
EIGHLLGLGHSS AIMYP+I RT+KV L DDI+GI+ LYG NP
Sbjct: 819 EIGHLLGLGHSSDLRAIMYPSIPPRTRKVNLAQDDIDGIRKLYGINP 959
>TC232889 weakly similar to UP|O65340 (O65340) Metalloproteinase , partial
(38%)
Length = 734
Score = 293 bits (750), Expect = 8e-80
Identities = 146/222 (65%), Positives = 168/222 (74%), Gaps = 8/222 (3%)
Frame = +1
Query: 155 VAHYSFFPGQPRWPEGTQTLTYAFDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYS 214
VAH++ FPG PRWPEGTQ LTYAF P L DA K VFA AF +W++VT++ F E SY
Sbjct: 1 VAHFTLFPGMPRWPEGTQELTYAFFPGNGLSDAVKGVFAAAFARWAEVTSLKFRETASYF 180
Query: 215 SSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAV 274
+DI+IGF+SGDHGDGE FDG LGTLAHAFSPT+GR HLD AEDWVV+GDVT S+L AV
Sbjct: 181 GADIRIGFFSGDHGDGEPFDGSLGTLAHAFSPTNGRFHLDAAEDWVVSGDVTRSALPTAV 360
Query: 275 DLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFTGT 334
DLESV VHEIGHLLGLGHSS+EEA+M+PTISSR KKV L DDIEGIQ LYGSNPNF G+
Sbjct: 361 DLESVAVHEIGHLLGLGHSSVEEAVMFPTISSRKKKVVLARDDIEGIQFLYGSNPNFNGS 540
Query: 335 TTATSRDRDSSFGGR--------HGVSLFLSLVFLGFGFLSL 368
T ++ +RD+S GGR GV L+ FL FL L
Sbjct: 541 TATSAPERDASDGGRCLTCVGPSSGVFALLTFAFLF*VFLLL 666
>CO985121
Length = 535
Score = 219 bits (557), Expect = 2e-57
Identities = 110/154 (71%), Positives = 127/154 (82%), Gaps = 6/154 (3%)
Frame = -2
Query: 217 DIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDL 276
DI+IGFY GDHGDGE FDGVLGTLAHA PT+G HLD AEDWV +GDVT++SLSNAVDL
Sbjct: 534 DIRIGFYGGDHGDGEPFDGVLGTLAHAXXPTNGMFHLDSAEDWVASGDVTKASLSNAVDL 355
Query: 277 ESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFTGTTT 336
ESV VHEIGHLLGLGHSS+E+AIMYPTI++RT+KVEL DDI+GIQ+LYGSNPNFTGT
Sbjct: 354 ESVAVHEIGHLLGLGHSSVEDAIMYPTITARTRKVELNEDDIQGIQVLYGSNPNFTGTPA 175
Query: 337 ATSRDRDSSFGG-RH-----GVSLFLSLVFLGFG 364
+SR+ D+S GG RH GV F L+F+ FG
Sbjct: 174 TSSRESDTSDGGVRHVGATCGV-FFSLLLFVVFG 76
>TC212939 weakly similar to UP|Q9ZUJ5 (Q9ZUJ5) T2K10.2 protein, partial (13%)
Length = 821
Score = 189 bits (481), Expect = 1e-48
Identities = 96/145 (66%), Positives = 112/145 (77%), Gaps = 6/145 (4%)
Frame = +1
Query: 1 MITQNQRYYYHIFYITLI---SITLTSVSARFFPDPSSMPAWNSSNAPP-GAWDGYKNFT 56
M T +Q+ Y + TLI S T VSAR FP+ SS+P WN+S+APP AW+ Y+NFT
Sbjct: 385 MSTSHQQRYLLFAFTTLIVFFSFNTTPVSARLFPNVSSIPRWNTSHAPPDSAWNAYRNFT 564
Query: 57 GCSRGKTYDGLSKLKNYFNHFGYIPN--GPPSNFTDDFDEALESAVRTYQKNFNLNITGE 114
GC GKTY+GLS LKNYF++FGYI N SNF+DDFD+ALE+AVR YQKNFNLNITGE
Sbjct: 565 GCRPGKTYNGLSNLKNYFHYFGYISNVSSKSSNFSDDFDDALEAAVRAYQKNFNLNITGE 744
Query: 115 LDDATMNYIVKPRCGVADIINGTTS 139
LDD TMN IVKPRCGVADIINGTT+
Sbjct: 745 LDDPTMNQIVKPRCGVADIINGTTT 819
>TC203322 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (74%)
Length = 1664
Score = 140 bits (353), Expect = 9e-34
Identities = 98/309 (31%), Positives = 159/309 (50%), Gaps = 6/309 (1%)
Frame = +2
Query: 66 GLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVK 125
GLS +K+Y + +GYI + P FTD FD+ + SA++TYQ NL +TG L+ + ++
Sbjct: 422 GLSVVKDYLSDYGYIESSGP--FTDSFDQEIISAIKTYQNFSNLQVTGGLNKQLIQQMLS 595
Query: 126 PRCGVADIINGTTSMNSGKFNSSSTNF-HTVAHYSFFPGQPRWPEGTQTLTYAFDPSENL 184
RCGV D+ + +N + N + A + +FP + LTY F P +
Sbjct: 596 IRCGVPDV--------NFDYNFTDDNISYPKAGHRWFPNR--------NLTYGFLPENQI 727
Query: 185 DDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAF 244
D +VF ++F +W++ + T+Y ++DI++GFY+ + E + G L
Sbjct: 728 PDNMTKVFRDSFARWAQASGTLSLRETTYDNADIQVGFYNFTNSRIEVYGGSTIFL-QPD 904
Query: 245 SPTDGRLHLDKAEDWVVNGD-VTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPT 303
S G + LD W++ + T S +DLE+V +H+IGHLLGL HS E+++MYP
Sbjct: 905 SSKKGVVLLDGNMGWLLPSENATLSKDDGVLDLETVAMHQIGHLLGLDHSHKEDSVMYPY 1084
Query: 304 I---SSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFGGRHGVSLFLSLVF 360
I S +KV+L + D I + + + + S D +S GG G+ + L F
Sbjct: 1085ILSSQSEQRKVQLSNSDKANIHLQFAKHD*DVTSLLPNSHD-SASHGG--GLDVLLVTTF 1255
Query: 361 -LGFGFLSL 368
LGF ++ L
Sbjct: 1256SLGFAYVLL 1282
>TC203267 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (61%)
Length = 1338
Score = 132 bits (331), Expect = 3e-31
Identities = 87/266 (32%), Positives = 144/266 (53%), Gaps = 3/266 (1%)
Frame = +3
Query: 66 GLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVK 125
GLS +K+Y + +GYI + P F + FD+ SA++TYQK NL +TG + + ++
Sbjct: 303 GLSVVKDYLSEYGYIESSRP--FNNSFDQETMSAIKTYQKFSNLPVTGVPNKQLIQQMLS 476
Query: 126 PRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLD 185
RCGV D+ F +T++ H +FP + LTY F P +
Sbjct: 477 LRCGVPDV------NFDYNFTDDNTSYPKAGH-RWFPNR--------NLTYGFLPENQIP 611
Query: 186 DATKQVFANAFNQWSKVT-TITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAF 244
D +VF ++F +W++ + T++ TE T+Y ++DI++GFY+ + E + G L L
Sbjct: 612 DNMTKVFRDSFARWAQASGTLSLTE-TTYDNADIQVGFYNFTNSRIEVYGGSLIFL-QPD 785
Query: 245 SPTDGRLHLDKAEDWVVNGD-VTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPT 303
S G + L+ W++ + T S +DLE+ +H+IGHLLGL HS E+++MYP
Sbjct: 786 SSKKGVVLLNGNMGWLLPSENATLSKDDGVLDLETAAMHQIGHLLGLDHSHKEDSVMYPY 965
Query: 304 I-SSRTKKVELESDDIEGIQMLYGSN 328
I SS+ +KV+L + D I + + +
Sbjct: 966 ILSSQQRKVQLSNSDKANIHLQFAKH 1043
>TC204855 UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2, complete
Length = 1307
Score = 127 bits (319), Expect = 8e-30
Identities = 106/323 (32%), Positives = 161/323 (49%), Gaps = 8/323 (2%)
Frame = +1
Query: 54 NFTGC--SRGKTYDGLSKLKNYFNHFGYIPN-GPPSNFTDDFDEALESAVRTYQKNFNLN 110
NFT S ++ +S +K+Y +++GYI + GP SN D E + SA++TYQ+ F+L
Sbjct: 232 NFTEIFSSEERSAPPVSLIKDYLSNYGYIESSGPLSNSMDQ--ETIISAIKTYQQYFSLQ 405
Query: 111 ITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEG 170
TG+L++ T+ + RCGV DI N F + ++ H RW
Sbjct: 406 PTGKLNNETLQQMSFLRCGVPDI---NIDYN---FTDDNMSYPKAGH--------RWFPH 543
Query: 171 TQTLTYAFDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHG-- 228
T LTY F P + +VF ++F +W++ + + T+Y ++DI++GFY+ +
Sbjct: 544 TN-LTYGFLPENQIPANMTKVFRDSFARWAQASGVLNLTETTYDNADIQVGFYNFTYLGI 720
Query: 229 DGEAFDGVLGTLAHAFSPTDGRLHLDKAED-WVVNGDVTESSLSNAV-DLESVVVHEIGH 286
D E + G L L S G + LD W + + S V DLES +HEIGH
Sbjct: 721 DIEVYGGSLIFL-QPDSTKKGVILLDGTNKLWALPSENGRLSWEEGVLDLESAAMHEIGH 897
Query: 287 LLGLGHSSIEEAIMYPTI-SSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSS 345
LLGL HS+ E+++MYP I S +KV+L D +Q + + + G
Sbjct: 898 LLGLDHSNKEDSVMYPCILPSHQRKVQLSKSDKTNVQHQFANVEDSAGHV---------- 1047
Query: 346 FGGRHGVSLFLSLVFLGFGFLSL 368
GR GVSL +L LGF +L L
Sbjct: 1048--GRLGVSLITTL-SLGFAYLLL 1107
>AW597906 weakly similar to PIR|T00643|T006 zinc metalloproteinase homolog
F3I6.6 - Arabidopsis thaliana, partial (11%)
Length = 279
Score = 126 bits (316), Expect = 2e-29
Identities = 61/79 (77%), Positives = 64/79 (80%), Gaps = 1/79 (1%)
Frame = -2
Query: 119 TMNYIVKPRCGVADIINGTTSMNSGKFNSS-STNFHTVAHYSFFPGQPRWPEGTQTLTYA 177
TMN IVKPRCGVADIINGTT+MNSGK N++ S FHTVAHYSFF GQPRWP GTQ LTYA
Sbjct: 239 TMNQIVKPRCGVADIINGTTTMNSGKTNTTDSPTFHTVAHYSFFDGQPRWPVGTQELTYA 60
Query: 178 FDPSENLDDATKQVFANAF 196
FDP LDD K VF NAF
Sbjct: 59 FDPDNALDDVIKTVFGNAF 3
>BU762711
Length = 428
Score = 105 bits (263), Expect = 2e-23
Identities = 52/89 (58%), Positives = 68/89 (75%)
Frame = +1
Query: 253 LDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKVE 312
LD AE W V+ + ES + AVDLESV HEIGH+LGLGHSS++EA+MYP++S R KKV+
Sbjct: 1 LDAAETWSVDFEREESRV--AVDLESVATHEIGHVLGLGHSSVKEAVMYPSLSPRRKKVD 174
Query: 313 LESDDIEGIQMLYGSNPNFTGTTTATSRD 341
L DD+ G+Q LYGSNPNFT ++ S++
Sbjct: 175 LRIDDVVGVQALYGSNPNFTFSSLLQSQN 261
>AW596048 weakly similar to GP|16901508|gb| matrix metalloproteinase MMP2
{Glycine max}, partial (16%)
Length = 417
Score = 86.3 bits (212), Expect = 2e-17
Identities = 51/142 (35%), Positives = 78/142 (54%), Gaps = 1/142 (0%)
Frame = +3
Query: 88 FTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNS 147
F DD D+A SA+ TYQ+ FNL ITG+L + T+ I PRCGV D +N ++ +
Sbjct: 24 FNDDLDQATVSAITTYQRFFNLKITGDLTNETLQQISLPRCGVPD-MNFDYDVSKDNVSW 200
Query: 148 SSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDDATKQVFANAFNQWS-KVTTIT 206
+ +H +FP + LTY F P+ + +VF +AF +W+ V +
Sbjct: 201 PMSRYHR----RWFP--------DRNLTYGFSPASKIPSNATKVFRDAFARWAGSVPGLN 344
Query: 207 FTEATSYSSSDIKIGFYSGDHG 228
TE +Y+S+D+K+GFY+ D G
Sbjct: 345 LTE-MNYNSADLKVGFYNLDEG 407
>TC203384 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (32%)
Length = 643
Score = 70.9 bits (172), Expect = 9e-13
Identities = 56/165 (33%), Positives = 88/165 (52%), Gaps = 11/165 (6%)
Frame = +1
Query: 215 SSDIKIGFYSGD--HGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGD-VTESSLS 271
++DI++GFY+ + E + G L L S G + +D W++ + T S
Sbjct: 4 NADIQVGFYNFTALSIEVEVYGGSLIFL-QPDSSKKGMVLMDGNMGWLLPSENATLSKDD 180
Query: 272 NAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTI-SSRTKKVELESDDIEGIQMLYGS--- 327
+DLE+V +H+IGHLLGL HS E+++MYP I SS+ +KV+L + D I + + S
Sbjct: 181 GVLDLETVAMHQIGHLLGLDHSHKEDSVMYPYILSSQQRKVKLSNSDKANIHLQFASHDS 360
Query: 328 ---NPNFTGTTTATSRDRDS-SFGGRHGVSLFLSLVFLGFGFLSL 368
+PN + + DS S GGR V L ++ LGF ++ L
Sbjct: 361 DMTSPNSHDSDLTSPNSHDSASHGGRLDV-LLVTTFSLGFAYVLL 492
>TC203453 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (29%)
Length = 622
Score = 66.2 bits (160), Expect = 2e-11
Identities = 47/133 (35%), Positives = 72/133 (53%), Gaps = 9/133 (6%)
Frame = +1
Query: 245 SPTDGRLHLDKAEDWVVNGD-VTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPT 303
S G + +D W++ + T S +DLE+V +H+IGHLLGL HS E+++MYP
Sbjct: 70 SSKKGVVLMDGNMGWLLPSENATLSKDDGVLDLETVAMHQIGHLLGLDHSHKEDSVMYPY 249
Query: 304 I-SSRTKKVELESDDIEGIQMLYGS------NPNFTGTTTATSRDRDS-SFGGRHGVSLF 355
I SS+ +KV+L + D I + + S +PN + + DS S GGR V L
Sbjct: 250 ILSSQQRKVKLSNSDKANIHLQFASHDSDLTSPNSHDSDLTSPNSHDSASHGGRLDV-LL 426
Query: 356 LSLVFLGFGFLSL 368
++ LGF ++ L
Sbjct: 427 VTTFSLGFAYVLL 465
>TC203366 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (18%)
Length = 576
Score = 64.3 bits (155), Expect = 8e-11
Identities = 30/68 (44%), Positives = 45/68 (66%)
Frame = +1
Query: 66 GLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVK 125
GLS +K+Y + +GYI + P F D FD+ + SA++TYQ NLN+TG+L+ + I+
Sbjct: 337 GLSVVKDYLSDYGYIESSGP--FNDSFDQEIISAIKTYQNFSNLNVTGDLNKQLIQQILS 510
Query: 126 PRCGVADI 133
RCGV D+
Sbjct: 511 IRCGVPDV 534
>TC203520 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (13%)
Length = 434
Score = 59.3 bits (142), Expect = 3e-09
Identities = 38/94 (40%), Positives = 58/94 (61%), Gaps = 1/94 (1%)
Frame = +2
Query: 276 LESVVVHEIGHLLGLGHSSIEEAIMYPTI-SSRTKKVELESDDIEGIQMLYGSNPNFTGT 334
LE+V +H+IGHLLGL HS E+++MYP I SS+ +KV+L + D I + + + +
Sbjct: 2 LETVAMHQIGHLLGLEHSPKEDSVMYPYILSSQQRKVKLSNSDKANIHLEFAKHD--SDL 175
Query: 335 TTATSRDRDSSFGGRHGVSLFLSLVFLGFGFLSL 368
T+ S D +S GGR V L ++ LGF ++ L
Sbjct: 176 TSPNSHD-SASHGGRLDV-LLVTTFSLGFAYVLL 271
>TC203588 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (14%)
Length = 514
Score = 45.8 bits (107), Expect = 3e-05
Identities = 22/51 (43%), Positives = 33/51 (64%)
Frame = +3
Query: 66 GLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELD 116
GL +K+Y + +GYI + P F D FD+ + SA++TYQ NL +TG L+
Sbjct: 345 GLYIVKDYLSDYGYIESSGP--FNDSFDQEIISAIKTYQNFSNLQVTGGLN 491
>TC204369 similar to GB|BAB09857.1|9759402|AB008268 phi-1-like protein
{Arabidopsis thaliana;} , partial (75%)
Length = 1276
Score = 33.5 bits (75), Expect = 0.16
Identities = 20/76 (26%), Positives = 33/76 (43%)
Frame = +3
Query: 130 VADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDDATK 189
+ D IN +S + S++T + T Y G P G Q L A+ +NL
Sbjct: 276 IVDFINSLSSAPNAPLPSTATWWKTTEKYKVGGGSPALVVGMQFLHPAYTLGKNLKGRDL 455
Query: 190 QVFANAFNQWSKVTTI 205
A+ FN+ + +T +
Sbjct: 456 LALASKFNEINSITVV 503
>BI423800 similar to GP|27902363|gb| polyphenol oxidase {Trifolium pratense},
partial (6%)
Length = 421
Score = 30.0 bits (66), Expect = 1.7
Identities = 16/37 (43%), Positives = 21/37 (56%)
Frame = +2
Query: 73 YFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNL 109
+F H +P G +FTDD D+ LES+ Y N NL
Sbjct: 77 FFAHHSNVPGGYRRDFTDD-DDWLESSFLFYDGNKNL 184
>TC220722 similar to UP|Q9LM55 (Q9LM55) F2E2.11, partial (28%)
Length = 776
Score = 30.0 bits (66), Expect = 1.7
Identities = 15/41 (36%), Positives = 22/41 (53%), Gaps = 4/41 (9%)
Frame = +1
Query: 176 YAFDPSENLDDATKQVFANAFNQW----SKVTTITFTEATS 212
YAFDPS ++D +V+ ++W KV FTE+ S
Sbjct: 133 YAFDPSNSVDSGRIKVYDQGEDEWKVVIGKVPVYDFTESES 255
>TC217490 similar to UP|Q9FNN8 (Q9FNN8) Cleft lip and palate associated
transmembrane protein-like, partial (60%)
Length = 1419
Score = 30.0 bits (66), Expect = 1.7
Identities = 16/41 (39%), Positives = 21/41 (51%)
Frame = +3
Query: 144 KFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENL 184
KFN + V H + P PE T+TLT + PSE+L
Sbjct: 450 KFNDFGSESSLVWHETNIPYAVWGPESTRTLTLKYHPSESL 572
>AI416519
Length = 274
Score = 29.6 bits (65), Expect = 2.3
Identities = 20/54 (37%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Frame = -2
Query: 111 ITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVA-HY--SFF 161
+TG +D + KP CG II G+ ++N+G S F A HY SFF
Sbjct: 192 VTGRRNDGG*DAAGKPCCGGGGIIKGSGNVNTGIQTKSQLQFPPFASHYFLSFF 31
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,516,078
Number of Sequences: 63676
Number of extensions: 241682
Number of successful extensions: 1371
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1352
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1353
length of query: 368
length of database: 12,639,632
effective HSP length: 99
effective length of query: 269
effective length of database: 6,335,708
effective search space: 1704305452
effective search space used: 1704305452
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144345.3


