

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
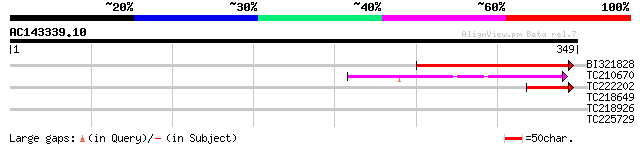
Query= AC143339.10 + phase: 0 /pseudo
(349 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI321828 118 5e-27
TC210670 weakly similar to UP|Q8DMU5 (Q8DMU5) Tll0016 protein, p... 49 3e-06
TC222202 weakly similar to GB|AAF68109.1|7715591|AC010793 F20B17... 49 4e-06
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 31 0.73
TC218926 similar to UP|CAZ_DROME (Q27294) RNA-binding protein ca... 28 8.1
TC225729 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precur... 28 8.1
>BI321828
Length = 367
Score = 118 bits (295), Expect = 5e-27
Identities = 51/97 (52%), Positives = 74/97 (75%)
Frame = +1
Query: 251 SMYKTIKYPQAWKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVA 310
+M+ T++ W P LYM+LSLAL++ EG FYWYTDSK GP+FSQE VG I++I +V
Sbjct: 1 AMWTTLRSEDVWGPCLYMYLSLALSLDIREGMFYWYTDSKGGPSFSQESVGFIFSISSVG 180
Query: 311 SIIGVLIYHKYLKNYTFRNLVFYAQLLYAISGLLDLM 347
+++G ++Y LK+Y FRNL+F+ QL+Y +SG+LDL+
Sbjct: 181 ALLGAILYQYALKDYAFRNLLFWTQLIYGLSGMLDLI 291
>TC210670 weakly similar to UP|Q8DMU5 (Q8DMU5) Tll0016 protein, partial (33%)
Length = 745
Score = 48.9 bits (115), Expect = 3e-06
Identities = 32/140 (22%), Positives = 71/140 (49%), Gaps = 5/140 (3%)
Frame = +3
Query: 209 GLMAVFPALTIVLGFVIYEKRTTGLHNEKK-----KGVVENVGTTIRSMYKTIKYPQAWK 263
G+ ++ P +T V+ ++ E+ G +E+ I ++ +++ +
Sbjct: 21 GVTSLLPLITSVVAVLVKEQPMIGTARGLNLLFSGPEFLESSKQRIIQLWGSVRQRSVFL 200
Query: 264 PSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLK 323
P+L++FL A + +++T + +G F+ EF+G + + ++AS++GV +Y+ +LK
Sbjct: 201 PTLFIFLWQA--TPQSDSAMFYFTTNSLG--FTPEFLGRVKLVTSIASLLGVGLYNGFLK 368
Query: 324 NYTFRNLVFYAQLLYAISGL 343
N R + F LL + G+
Sbjct: 369 NVPLRKVFFATTLLGSTLGM 428
>TC222202 weakly similar to GB|AAF68109.1|7715591|AC010793 F20B17.13
{Arabidopsis thaliana;} , partial (25%)
Length = 456
Score = 48.5 bits (114), Expect = 4e-06
Identities = 22/29 (75%), Positives = 26/29 (88%)
Frame = +2
Query: 319 HKYLKNYTFRNLVFYAQLLYAISGLLDLM 347
HK LK+Y FR+LVFYAQLLY ISG+LDL+
Sbjct: 2 HKALKDYPFRDLVFYAQLLYGISGVLDLI 88
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 31.2 bits (69), Expect = 0.73
Identities = 16/43 (37%), Positives = 26/43 (60%)
Frame = +2
Query: 171 GLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAV 213
GL+ SL G S GA++G +ASG ++G +GSL + ++
Sbjct: 272 GLSVSEFSLFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIASI 400
>TC218926 similar to UP|CAZ_DROME (Q27294) RNA-binding protein cabeza
(Sarcoma-associated RNA-binding fly homolog) (P19),
partial (5%)
Length = 906
Score = 27.7 bits (60), Expect = 8.1
Identities = 23/88 (26%), Positives = 36/88 (40%), Gaps = 8/88 (9%)
Frame = +3
Query: 112 RPYFVVAGVVGTVSATIVAVGG--EVSVVAALMCF------VGVSASLAIADVTIDACIA 163
RP+ A V SA +VAV + AL F + + SL++AD + +
Sbjct: 93 RPFTAFAAAVAGASAAVVAVSSSDRSFLRNALNSFFSSDLSMPLWGSLSLADSGVSVVES 272
Query: 164 RNSIEVPGLAPDLQSLCGFCSGSGALLG 191
+ P + Q LCG ++LG
Sbjct: 273 KTGTSFPSVLDSSQKLCGIGLRKKSILG 356
>TC225729 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(82%)
Length = 732
Score = 27.7 bits (60), Expect = 8.1
Identities = 23/70 (32%), Positives = 32/70 (44%)
Frame = -1
Query: 202 LGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQA 261
L P+GSL + AL ++LG V+ + G K + E GT
Sbjct: 189 LAPKGSLAYQPL--ALVLLLGGVLC*RPREG------KNIGEGSGTW------------Q 70
Query: 262 WKPSLYMFLS 271
WKPSL++FLS
Sbjct: 69 WKPSLFLFLS 40
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.141 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,865,124
Number of Sequences: 63676
Number of extensions: 222093
Number of successful extensions: 1475
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 1467
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1475
length of query: 349
length of database: 12,639,632
effective HSP length: 98
effective length of query: 251
effective length of database: 6,399,384
effective search space: 1606245384
effective search space used: 1606245384
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC143339.10


