

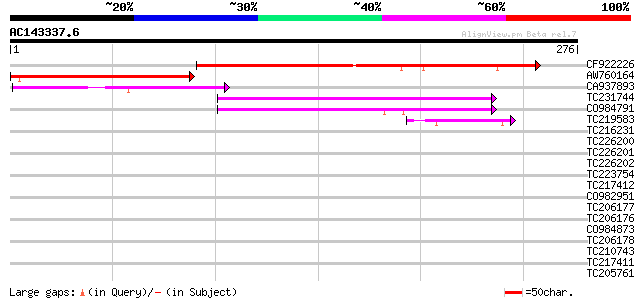
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.6 + phase: 0
(276 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922226 140 8e-34
AW760164 similar to GP|11994422|dbj oxidoreductase short-chain ... 93 1e-19
CA937893 similar to GP|20805072|dbj retrovirus-related pol polyp... 59 2e-09
TC231744 44 1e-04
CO984791 43 2e-04
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 42 3e-04
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 39 0.002
TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 38 0.006
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 38 0.006
TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 38 0.006
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 37 0.010
TC217412 similar to UP|Q6L724 (Q6L724) ATP-dependent RNA helicas... 36 0.017
CO982951 36 0.017
TC206177 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 36 0.017
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 35 0.037
CO984873 34 0.082
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 33 0.14
TC210743 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1,... 32 0.24
TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%) 32 0.24
TC205761 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%) 31 0.70
>CF922226
Length = 667
Score = 140 bits (352), Expect = 8e-34
Identities = 79/191 (41%), Positives = 117/191 (60%), Gaps = 24/191 (12%)
Frame = -3
Query: 92 MTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCAL 151
MTKSL +R + KQ LYSF+M E +++ EQL FNK++ DLENI+V ++DED+A+LLLC L
Sbjct: 665 MTKSLVNRLYXKQSLYSFKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYL 486
Query: 152 PKSFESFKDTMLYGKEGTVTLEEVQAALRTKDLTKSKD-LTHEHGEGLSV---------- 200
PKS+ FK+T+L+G++ +V+L+EVQ AL +K+L + K+ + GEGL+
Sbjct: 485 PKSYSHFKETLLFGRD-SVSLDEVQTALNSKELNERKEKKSSASGEGLTARGKTFKKDSE 309
Query: 201 -TRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP------------KINGNSAQIVS 247
+ + GN + C++C K GH +K CP K +GN+A +
Sbjct: 308 FDKKKQKPENQKNGEGNIFKIRCYHCKKEGHTRKVCPERQKNGGSNNRKKDSGNAAIVQD 129
Query: 248 EGYEDAGALMV 258
+GYE A ALMV
Sbjct: 128 DGYESAEALMV 96
>AW760164 similar to GP|11994422|dbj oxidoreductase short-chain
dehydrogenase/reductase family-like protein {Arabidopsis
thaliana}, partial (9%)
Length = 428
Score = 92.8 bits (229), Expect = 1e-19
Identities = 48/91 (52%), Positives = 66/91 (71%), Gaps = 1/91 (1%)
Frame = +3
Query: 1 MGS-KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKAR 59
MGS K+++EKFTG NDFGL +KM A+L+QQ +AL GE L M+ +K ++ KA
Sbjct: 75 MGSAKYEVEKFTGQNDFGLC*LKMRALLVQQGLVEALDGEIKLEKMMADGDKKALLQKAY 254
Query: 60 SAVVLCLGDKVLREVAKEATAASMWAKLESL 90
+A++L LGDKVLR+V+KE TA +W+KLE L
Sbjct: 255 NAIILSLGDKVLRQVSKETTAVGVWSKLEVL 347
>CA937893 similar to GP|20805072|dbj retrovirus-related pol polyprotein from
transposon TNT 1-94-like, partial (7%)
Length = 412
Score = 59.3 bits (142), Expect = 2e-09
Identities = 35/112 (31%), Positives = 61/112 (54%), Gaps = 6/112 (5%)
Frame = -2
Query: 2 GSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVD----- 56
G+K+++ KF G +F LW+ +++ +L Q K L+ S++ T+ +D
Sbjct: 315 GAKFEVGKFDGTGNFRLWQKRVKDLLA*QGLLKVLRD--------SKSNNTEALDWEEL* 160
Query: 57 -KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLY 107
+ + + LCL D+ L + + A +W KLES YM KSL ++ +L Q+LY
Sbjct: 159 ERTATTIRLCLVDEFLYHMMELAFPGEVWKKLESQYMLKSLTNKLYLMQKLY 4
>TC231744
Length = 794
Score = 43.5 bits (101), Expect = 1e-04
Identities = 30/136 (22%), Positives = 57/136 (41%)
Frame = +2
Query: 102 LKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDT 161
L +L S + I E + E + + L++++++L ++ L+L +LP F FK +
Sbjct: 41 LLDKLISMKYKGKGNIREYIMEISNLASKLKSLKLELGEDLFVHLVLISLPAHFGQFKVS 220
Query: 162 MLYGKEGTVTLEEVQAALRTKDLTKSKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFE 221
K+ E + ++ ++ H + T+ ++K F
Sbjct: 221 YNTQKDKWSLNELISHCVQEEERL*RDRTESAHNKKRKKTKDVAEKTS*QKKQQKDEEFT 400
Query: 222 CFNCHKMGHFKKDCPK 237
C+ C K H KK CPK
Sbjct: 401 CYFCKKSRHMKKKCPK 448
>CO984791
Length = 672
Score = 42.7 bits (99), Expect = 2e-04
Identities = 34/142 (23%), Positives = 63/142 (43%), Gaps = 6/142 (4%)
Frame = -3
Query: 102 LKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDT 161
L +L S + I E + E + + L++++++L ++ L+L +LP F FK +
Sbjct: 607 LLAKLISMKYKGKGNIREYIIEMSNLASKLKSLKLELGEDLLMYLVLISLPAHFGQFKVS 428
Query: 162 MLYGKEGTVTLEEVQAALRT-----KDLTKSKDL-THEHGEGLSVTRGNGGGRGNRRKSG 215
K+ E + ++ +D T+S L + + T+G G ++K
Sbjct: 427 YNT*KDKWSLNELISHCVQEEESL*RDRTESAHLISTSQNKKWKKTKGAA*GTS*QKKQN 248
Query: 216 NKSRFECFNCHKMGHFKKDCPK 237
F C+ K GH KK+C K
Sbjct: 247 KDEEFACYFYKKSGHMKKECLK 182
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 42.0 bits (97), Expect = 3e-04
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Frame = +2
Query: 194 HGEGLSVTRGNGG--GRGNRRKSGNKSRFECFNCHKMGHFKKDCPKI-NGNSAQIV 246
HG+G G GG GR RR G CFNC + GHF ++CP + GN + V
Sbjct: 470 HGQG-----GGGGDDGRNRRRGGGGGGGGGCFNCGEEGHFARECPNVGKGNE*ECV 622
Score = 37.7 bits (86), Expect = 0.006
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Frame = +2
Query: 164 YGKEGTVTLEEVQAALRTKDLTKSKDLTHEHGEGLS-VTRGNGGGRGNRRKSGNKSRFEC 222
YG +G +V +A+R++ + G G G G GRG R+ G EC
Sbjct: 257 YGDDGRTMAVDVTSAVRSRLPGGFRGGGGGRGRGGGRYGGGEGRGRGFGRRGGGP---EC 427
Query: 223 FNCHKMGHFKKDC 235
+NC ++GH +DC
Sbjct: 428 YNCGRIGHLARDC 466
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 39.3 bits (90), Expect = 0.002
Identities = 15/32 (46%), Positives = 18/32 (55%)
Frame = +3
Query: 205 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP 236
GGG G R G C+NC + GHF +DCP
Sbjct: 279 GGGGGGRYGGGGGGGGSCYNCGESGHFARDCP 374
Score = 29.3 bits (64), Expect = 2.0
Identities = 13/42 (30%), Positives = 18/42 (41%)
Frame = +3
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQ 244
G GGG G G C+ C + GH +DC + G +
Sbjct: 183 GGGGGYGGGGGGGGGG---CYKCGETGHIARDCSQGGGGGGR 299
>TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (89%)
Length = 936
Score = 37.7 bits (86), Expect = 0.006
Identities = 19/44 (43%), Positives = 28/44 (63%)
Frame = +2
Query: 201 TRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQ 244
+RG GGGRG R SG S +C+ C + GHF ++C ++ G S +
Sbjct: 284 SRGGGGGRGGR--SGG-SDLKCYECGEPGHFAREC-RMRGGSGR 403
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 37.7 bits (86), Expect = 0.006
Identities = 19/44 (43%), Positives = 28/44 (63%)
Frame = +1
Query: 201 TRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQ 244
+RG GGGRG R SG S +C+ C + GHF ++C ++ G S +
Sbjct: 193 SRGGGGGRGGR--SGG-SDLKCYECGEPGHFAREC-RMRGGSGR 312
>TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 1003
Score = 37.7 bits (86), Expect = 0.006
Identities = 19/44 (43%), Positives = 28/44 (63%)
Frame = +3
Query: 201 TRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQ 244
+RG GGGRG R SG S +C+ C + GHF ++C ++ G S +
Sbjct: 780 SRGGGGGRGGR--SGG-SDLKCYECGEPGHFAREC-RMRGGSGR 899
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 37.0 bits (84), Expect = 0.010
Identities = 18/38 (47%), Positives = 22/38 (57%)
Frame = +2
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKING 240
G GGG G G KS CFNC K GHF ++C + +G
Sbjct: 380 GGGGGSGG---GGGKST--CFNCGKPGHFARECVEASG 478
Score = 32.3 bits (72), Expect = 0.24
Identities = 18/47 (38%), Positives = 20/47 (42%)
Frame = +2
Query: 195 GEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGN 241
G G SV G GGG G+ CF C GH +DC GN
Sbjct: 182 GGGGSVGIGGGGGGGS-----------CFRCGGFGHMARDCATGKGN 289
Score = 32.0 bits (71), Expect = 0.31
Identities = 17/46 (36%), Positives = 19/46 (40%)
Frame = +2
Query: 204 NGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQIVSEG 249
N GG G G CFNC GH +DC + G S I G
Sbjct: 98 NSGGGGGGSGGG------CFNCGGFGHLARDCVRGGGGSVGIGGGG 217
>TC217412 similar to UP|Q6L724 (Q6L724) ATP-dependent RNA helicase, partial
(3%)
Length = 868
Score = 36.2 bits (82), Expect = 0.017
Identities = 14/35 (40%), Positives = 20/35 (57%)
Frame = +3
Query: 205 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKIN 239
GG R +R S ++ CFNC + GH DCP ++
Sbjct: 261 GGRRSSRPSSSDRFGGTCFNCGESGHRASDCPNVS 365
>CO982951
Length = 757
Score = 36.2 bits (82), Expect = 0.017
Identities = 16/50 (32%), Positives = 26/50 (52%)
Frame = -3
Query: 188 KDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPK 237
K +H +G + N + ++K+ ++ C C K GHF+KDCPK
Sbjct: 320 KKFVKKHDKGKGPLKINNNSKQIQKKTSKRNN--CHFCGKSGHFQKDCPK 177
>TC206177 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (22%)
Length = 406
Score = 36.2 bits (82), Expect = 0.017
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = +1
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP 236
G GGGR G C++C + GHF +DCP
Sbjct: 49 GGGGGRYGGGGGGGGGGGSCYSCGESGHFARDCP 150
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 35.0 bits (79), Expect = 0.037
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = +3
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP 236
G GGGR G C++C + GHF +DCP
Sbjct: 534 GGGGGRYVGGGGGGGGGGSCYSCGESGHFARDCP 635
Score = 32.0 bits (71), Expect = 0.31
Identities = 13/33 (39%), Positives = 17/33 (51%)
Frame = +3
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDC 235
G GGG G + G C+NC + GH +DC
Sbjct: 384 GGGGGYGGGGRGGGGGA--CYNCGESGHLARDC 476
>CO984873
Length = 754
Score = 33.9 bits (76), Expect = 0.082
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 2/57 (3%)
Frame = -2
Query: 217 KSRFECFNCHKMGHFKKDCPKING--NSAQIVSEGYEDAGALMVWCCLEEEKGDVSH 271
K+ +CF C+K GH KKDC K N I + + C E K +V+H
Sbjct: 372 KNESKCFFCNKKGHIKKDCSKFKSWLNKKNIPFD----------FVCYESNKVNVNH 232
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 33.1 bits (74), Expect = 0.14
Identities = 15/34 (44%), Positives = 18/34 (52%), Gaps = 1/34 (2%)
Frame = +2
Query: 203 GNGGGRGNRRKSGNKSR-FECFNCHKMGHFKKDC 235
G GGG G R G R C NC ++GH +DC
Sbjct: 782 GGGGGGGARGGGGGGYRDVVCRNCQQLGHMSRDC 883
Score = 28.5 bits (62), Expect = 3.5
Identities = 9/15 (60%), Positives = 10/15 (66%)
Frame = +2
Query: 222 CFNCHKMGHFKKDCP 236
C NC K GH +DCP
Sbjct: 650 CNNCRKTGHLARDCP 694
Score = 27.3 bits (59), Expect = 7.7
Identities = 7/17 (41%), Positives = 12/17 (70%)
Frame = +2
Query: 222 CFNCHKMGHFKKDCPKI 238
C NC + GH+ ++CP +
Sbjct: 344 CKNCKRPGHYARECPNV 394
>TC210743 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1, partial
(53%)
Length = 452
Score = 32.3 bits (72), Expect = 0.24
Identities = 24/82 (29%), Positives = 32/82 (38%)
Frame = +3
Query: 150 ALPKSFESFKDTMLYGKEGTVTLEEVQAALRTKDLTKSKDLTHEHGEGLSVTRGNGGGRG 209
A+ ++ ++ L G+ TV + Q D D E G G GGGRG
Sbjct: 240 AMDEAIDAMNGMDLDGRTITVDRAQPQQGSTRDD----GDRYRERGRDRDRDHGGGGGRG 407
Query: 210 NRRKSGNKSRFECFNCHKMGHF 231
+ ECF C K GHF
Sbjct: 408 SNGG-------ECFKCGKPGHF 452
>TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%)
Length = 1275
Score = 32.3 bits (72), Expect = 0.24
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = +3
Query: 205 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP 236
G R +R S ++ CFNC + GH DCP
Sbjct: 765 GSRRSSRPSSSDRFGGACFNCGESGHRASDCP 860
>TC205761 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%)
Length = 1023
Score = 30.8 bits (68), Expect = 0.70
Identities = 13/26 (50%), Positives = 16/26 (61%)
Frame = -1
Query: 200 VTRGNGGGRGNRRKSGNKSRFECFNC 225
V R N GGRGN + GN++R C C
Sbjct: 228 VQRSNRGGRGNGDRHGNETRRTCLFC 151
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,875,766
Number of Sequences: 63676
Number of extensions: 123064
Number of successful extensions: 888
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 843
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 882
length of query: 276
length of database: 12,639,632
effective HSP length: 96
effective length of query: 180
effective length of database: 6,526,736
effective search space: 1174812480
effective search space used: 1174812480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC143337.6


