

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142498.7 + phase: 0
(1705 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
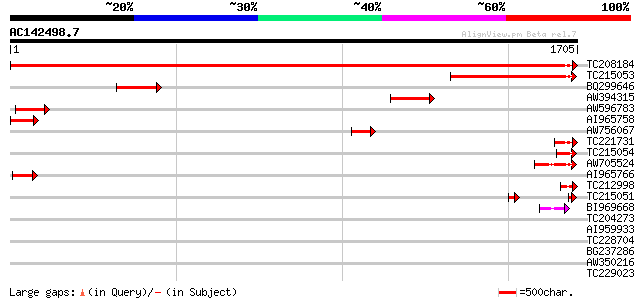
Score E
Sequences producing significant alignments: (bits) Value
TC208184 UP|Q39834 (Q39834) Clathrin heavy chain, complete 3182 0.0
TC215053 homologue to UP|Q39834 (Q39834) Clathrin heavy chain, p... 719 0.0
BQ299646 homologue to PIR|T06779|T0 clathrin heavy chain - soybe... 261 2e-69
AW394315 homologue to PIR|T06779|T0 clathrin heavy chain - soybe... 249 7e-66
AW596783 similar to PIR|T06779|T0 clathrin heavy chain - soybean... 186 1e-46
AI965758 homologue to PIR|T06779|T06 clathrin heavy chain - soyb... 159 8e-39
AW756067 homologue to PIR|T06779|T06 clathrin heavy chain - soyb... 131 3e-30
TC221731 UP|Q39834 (Q39834) Clathrin heavy chain, partial (4%) 117 4e-26
TC215054 homologue to UP|Q39834 (Q39834) Clathrin heavy chain, p... 112 1e-24
AW705524 112 2e-24
AI965766 85 3e-16
TC212998 similar to UP|Q8L3R8 (Q8L3R8) AT3g08530/T8G24_1, partia... 77 8e-14
TC215051 homologue to UP|Q39834 (Q39834) Clathrin heavy chain, p... 72 3e-12
BI969668 similar to SP|P49951|CLH_ Clathrin heavy chain. [Bovine... 45 4e-04
TC204273 similar to UP|Q8W2C2 (Q8W2C2) Phosphoenolpyruvate carbo... 37 0.057
AI959933 weakly similar to GP|27461187|gb type IV collagen alpha... 36 0.17
TC228704 similar to UP|Q9FJ12 (Q9FJ12) Arabidopsis thaliana geno... 36 0.17
BG237286 similar to PIR|T10586|T105 small nuclear ribonucleoprot... 36 0.17
AW350216 similar to PIR|T10586|T105 small nuclear ribonucleoprot... 36 0.17
TC229023 35 0.22
>TC208184 UP|Q39834 (Q39834) Clathrin heavy chain, complete
Length = 5624
Score = 3182 bits (8251), Expect = 0.0
Identities = 1610/1705 (94%), Positives = 1646/1705 (96%), Gaps = 2/1705 (0%)
Frame = +3
Query: 3 AAANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPNQP 62
AAANAPI MRE LT P+IGINPQ ITFTHVTMESDKYI VRETAPQ+SVVI+DMNMPNQP
Sbjct: 87 AAANAPIAMRETLTFPTIGINPQFITFTHVTMESDKYISVRETAPQDSVVIIDMNMPNQP 266
Query: 63 LRRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWIS 122
LRRPITADSALMNPNSR LALK QLQGTTQDHLQIFNIE K+KMKSYQ+PEQV FWKWI+
Sbjct: 267 LRRPITADSALMNPNSRSLALKTQLQGTTQDHLQIFNIEFKSKMKSYQIPEQVSFWKWIT 446
Query: 123 PKLLGLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSP 182
PKLLG+VTQTSVYHWSIEGDSEPVKMFERTANL NNQIINYRCDPTEKWLVLIGI GSP
Sbjct: 447 PKLLGIVTQTSVYHWSIEGDSEPVKMFERTANLPNNQIINYRCDPTEKWLVLIGILHGSP 626
Query: 183 ERPQLVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISK 242
ERPQLVKG MQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTL FAT NAGQ+I+K
Sbjct: 627 ERPQLVKGRMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLFLFATNP-NAGQIITK 803
Query: 243 LHVIELGAQPGKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLE 302
LHVIELGAQPG + + P F+DDFPV+MQISHKY+LIYVITKLGLLFVYDLE
Sbjct: 804 LHVIELGAQPGNHHLPRNKQIFSSPQIFSDDFPVAMQISHKYNLIYVITKLGLLFVYDLE 983
Query: 303 TATAVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELA 362
TATAVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELA
Sbjct: 984 TATAVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELA 1163
Query: 363 VNLAKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQA 422
VNLAKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQA
Sbjct: 1164 VNLAKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQA 1343
Query: 423 GQTPPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVK 482
GQTPPLLQYFGTLLTRGKLNA ESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVK
Sbjct: 1344 GQTPPLLQYFGTLLTRGKLNALESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVK 1523
Query: 483 TVDNDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQG 542
TVDNDLALKIYIKARATPKVVAAFAE+REFDKILIYSKQVGYTPDYLFLLQ+ILRTDPQG
Sbjct: 1524 TVDNDLALKIYIKARATPKVVAAFAERREFDKILIYSKQVGYTPDYLFLLQTILRTDPQG 1703
Query: 543 AVNFALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEI 602
AVNFALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHG+LQTKVLEI
Sbjct: 1704 AVNFALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGYLQTKVLEI 1883
Query: 603 NLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIE 662
NLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIE
Sbjct: 1884 NLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIE 2063
Query: 663 PQALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRS 722
PQ+LVEFFGTLS+EWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIK+FEQFRS
Sbjct: 2064 PQSLVEFFGTLSREWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKLFEQFRS 2243
Query: 723 YEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAK 782
YEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAK
Sbjct: 2244 YEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAK 2423
Query: 783 LPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPED 842
LPDARPLINVCDRFGFVPDLTHYLYT+NMLRYIEGYVQKVNPGNAPLVVGQLLDDECPED
Sbjct: 2424 LPDARPLINVCDRFGFVPDLTHYLYTNNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPED 2603
Query: 843 FIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSN 902
FIKGLILSVRSLLPVEPLV ECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSN
Sbjct: 2604 FIKGLILSVRSLLPVEPLVEECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSN 2783
Query: 903 NNPEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYV 962
NNPEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRG+CDDELINVTNKNSLFKLQARYV
Sbjct: 2784 NNPEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGLCDDELINVTNKNSLFKLQARYV 2963
Query: 963 VERMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIEL 1022
VERMD DLWEKVLNPDN YRRQLIDQVVSTALPESKSPEQVSA+VKAFMTADLPHELIEL
Sbjct: 2964 VERMDGDLWEKVLNPDNAYRRQLIDQVVSTALPESKSPEQVSAAVKAFMTADLPHELIEL 3143
Query: 1023 LEKIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEE 1082
LEKIVLQNSAFSGNFNLQNLLILTAIKAD SRVMDY+NRLDNFDGP VGE+AVEA+LYEE
Sbjct: 3144 LEKIVLQNSAFSGNFNLQNLLILTAIKADPSRVMDYINRLDNFDGPAVGEMAVEAQLYEE 3323
Query: 1083 AFAIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIES 1142
AFAIFKKFNLNVQAVNVLLD+IHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIES
Sbjct: 3324 AFAIFKKFNLNVQAVNVLLDHIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIES 3503
Query: 1143 FIRADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIE 1202
FIRADDATQFLDVIRAA+DGNAY DLVRYLLMVR KTKEPKVDSELIYAYAKIDRLSDIE
Sbjct: 3504 FIRADDATQFLDVIRAAEDGNAYHDLVRYLLMVRHKTKEPKVDSELIYAYAKIDRLSDIE 3683
Query: 1203 EFILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKA 1262
EFILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKA
Sbjct: 3684 EFILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKA 3863
Query: 1263 NSAKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGL 1322
NSAKTWKEVCFACVD EEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGL
Sbjct: 3864 NSAKTWKEVCFACVDAEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGL 4043
Query: 1323 GLERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQ 1382
GLERAHMGIFTELGVLYARYR EKLMEHIKLFATR NIPKLIRACDEQQHWKELTYLYIQ
Sbjct: 4044 GLERAHMGIFTELGVLYARYRHEKLMEHIKLFATRPNIPKLIRACDEQQHWKELTYLYIQ 4223
Query: 1383 YDEFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLA 1442
YDEFDNAATTIMNHSPEAWDHMQ KDV+ KVANVELYYKAVHFYLQEHPDLINDVLNVLA
Sbjct: 4224 YDEFDNAATTIMNHSPEAWDHMQLKDVVVKVANVELYYKAVHFYLQEHPDLINDVLNVLA 4403
Query: 1443 LRVDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLH 1502
LRVD ARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLH
Sbjct: 4404 LRVDQARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLH 4583
Query: 1503 DNFDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIAL-SKKDNLYKDAMETASQSGE 1561
DNFDQIGLAQKIE HELLEMRRVAAYIYKKAGRWKQSI L KKD LYKDAMETASQSG+
Sbjct: 4584 DNFDQIGLAQKIETHELLEMRRVAAYIYKKAGRWKQSIELCHKKDTLYKDAMETASQSGD 4763
Query: 1562 RELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREY 1621
RELAEELLVYFIDQGKKECFASCLFVCYDLIR D+ALELAWM+NMIDFAFPYLLQFIREY
Sbjct: 4764 RELAEELLVYFIDQGKKECFASCLFVCYDLIRADIALELAWMNNMIDFAFPYLLQFIREY 4943
Query: 1622 TGKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYA 1681
TGKVDELVK KIE+QN+VKAKEQEEKEVIAQQNMYAQLLPLALPAPPMP GMGGG+A
Sbjct: 4944 TGKVDELVKDKIEAQNQVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMP----GMGGGFA 5111
Query: 1682 PPPPMGGGMGMPPMPPFGM-PMGGY 1705
PPPPM GG+GMPPMPPFGM PMG Y
Sbjct: 5112 PPPPM-GGLGMPPMPPFGMPPMGSY 5183
Score = 30.4 bits (67), Expect = 7.0
Identities = 15/32 (46%), Positives = 17/32 (52%)
Frame = -2
Query: 1668 PMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFG 1699
PM G+ G GG PP GGG PP+P G
Sbjct: 5176 PMGGIPKGGIGGIPNPPIGGGGAKPPPIPGIG 5081
>TC215053 homologue to UP|Q39834 (Q39834) Clathrin heavy chain, partial (22%)
Length = 1446
Score = 719 bits (1855), Expect = 0.0
Identities = 355/380 (93%), Positives = 366/380 (95%), Gaps = 1/380 (0%)
Frame = +2
Query: 1326 RAH-MGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQYD 1384
R H MGIFTELGVLYARYRPEKLMEHIKLF+TRLNIPKLIRACDEQQHWKELTYLYIQYD
Sbjct: 2 RGHDMGIFTELGVLYARYRPEKLMEHIKLFSTRLNIPKLIRACDEQQHWKELTYLYIQYD 181
Query: 1385 EFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLALR 1444
EFDNAATTIMNHSPEAWDHMQFKDV+ KVANVELYYKAVHFYL+EHPDLINDVLNVLALR
Sbjct: 182 EFDNAATTIMNHSPEAWDHMQFKDVVVKVANVELYYKAVHFYLEEHPDLINDVLNVLALR 361
Query: 1445 VDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDN 1504
VDHARVVDIMRKAGHLRLVKPYM+AVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDN
Sbjct: 362 VDHARVVDIMRKAGHLRLVKPYMIAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDN 541
Query: 1505 FDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGEREL 1564
FDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGEREL
Sbjct: 542 FDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGEREL 721
Query: 1565 AEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGK 1624
AEELLVYFIDQGKKECFASCLFVCYDLIR D+ LELAWMHNMIDFAFPYLLQFIREYTGK
Sbjct: 722 AEELLVYFIDQGKKECFASCLFVCYDLIRADIVLELAWMHNMIDFAFPYLLQFIREYTGK 901
Query: 1625 VDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYAPPP 1684
VDELVK KIE+QNEVKAKEQEEK+V+AQQNMYAQLLPLALPAPPMPGM GG+APPP
Sbjct: 902 VDELVKDKIEAQNEVKAKEQEEKDVVAQQNMYAQLLPLALPAPPMPGM-----GGFAPPP 1066
Query: 1685 PMGGGMGMPPMPPFGMPMGG 1704
PM GG+GMPPMPP+GMP G
Sbjct: 1067 PM-GGLGMPPMPPYGMPPMG 1123
Score = 33.5 bits (75), Expect = 0.82
Identities = 17/33 (51%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Frame = -1
Query: 1668 PMPGMG-GGMGGGYAPPPPMGGGMGMPPMPPFG 1699
PM G+ GG+GG P PP+GGG PP+P G
Sbjct: 1122 PMGGIP*GGIGG--IPSPPIGGGGAKPPIPGIG 1030
>BQ299646 homologue to PIR|T06779|T0 clathrin heavy chain - soybean, partial
(8%)
Length = 409
Score = 261 bits (667), Expect = 2e-69
Identities = 135/136 (99%), Positives = 136/136 (99%)
Frame = +2
Query: 320 TSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVNLAKRGNLPGAEKLVV 379
TSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAV+LAKRGNLPGAEKLVV
Sbjct: 2 TSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVSLAKRGNLPGAEKLVV 181
Query: 380 ERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQYFGTLLTRG 439
ERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQYFGTLLTRG
Sbjct: 182 ERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQYFGTLLTRG 361
Query: 440 KLNAFESLELSRLVVN 455
KLNAFESLELSRLVVN
Sbjct: 362 KLNAFESLELSRLVVN 409
>AW394315 homologue to PIR|T06779|T0 clathrin heavy chain - soybean, partial
(7%)
Length = 403
Score = 249 bits (636), Expect = 7e-66
Identities = 124/134 (92%), Positives = 129/134 (95%)
Frame = +2
Query: 1144 IRADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIEE 1203
IRADDATQFLDVIRAA+DGN Y DLV+YLLMVR K KEPKVDSELIYAYAKIDRLSDIEE
Sbjct: 2 IRADDATQFLDVIRAAEDGNVYHDLVKYLLMVRGKAKEPKVDSELIYAYAKIDRLSDIEE 181
Query: 1204 FILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKAN 1263
FILMPNVANLQNVGDRLYDE LYEAAKIIFAFISNWAKLA+TLVKL+QFQGAVDAARKAN
Sbjct: 182 FILMPNVANLQNVGDRLYDEALYEAAKIIFAFISNWAKLAITLVKLQQFQGAVDAARKAN 361
Query: 1264 SAKTWKEVCFACVD 1277
S+KTWKEVCFACVD
Sbjct: 362 SSKTWKEVCFACVD 403
>AW596783 similar to PIR|T06779|T0 clathrin heavy chain - soybean, partial
(5%)
Length = 301
Score = 186 bits (471), Expect = 1e-46
Identities = 89/100 (89%), Positives = 95/100 (95%)
Frame = +2
Query: 19 SIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPNQPLRRPITADSALMNPNS 78
+IGINPQ IT THVTMESDKYICVRETAPQN+V I+DMNMPNQPLRRPITADSALMNPNS
Sbjct: 2 TIGINPQFITCTHVTMESDKYICVRETAPQNTVAIIDMNMPNQPLRRPITADSALMNPNS 181
Query: 79 RILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFW 118
RILALKAQLQGTT DHLQIFNIE++ KMKSYQMP+QVVFW
Sbjct: 182 RILALKAQLQGTTPDHLQIFNIEMRRKMKSYQMPDQVVFW 301
>AI965758 homologue to PIR|T06779|T06 clathrin heavy chain - soybean, partial
(4%)
Length = 411
Score = 159 bits (403), Expect = 8e-39
Identities = 80/85 (94%), Positives = 81/85 (95%)
Frame = +1
Query: 3 AAANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPNQP 62
AAANAPI MREALTLPSIGINPQ ITFTHVTMESDKYICVRETAPQNSVVI+DMNMPNQP
Sbjct: 124 AAANAPIAMREALTLPSIGINPQFITFTHVTMESDKYICVRETAPQNSVVIIDMNMPNQP 303
Query: 63 LRRPITADSALMNPNSRILALKAQL 87
LRRPITADSALMNPNSRILALK L
Sbjct: 304 LRRPITADSALMNPNSRILALKGWL 378
>AW756067 homologue to PIR|T06779|T06 clathrin heavy chain - soybean, partial
(4%)
Length = 214
Score = 131 bits (329), Expect = 3e-30
Identities = 66/71 (92%), Positives = 68/71 (94%)
Frame = +2
Query: 1029 QNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEEAFAIFK 1088
QNSAFSGNFNLQNLLILTAIKAD SRVMDYVNRLDNFDGP VG +AVEA+LYEEAFAIFK
Sbjct: 2 QNSAFSGNFNLQNLLILTAIKADPSRVMDYVNRLDNFDGPAVGGMAVEAQLYEEAFAIFK 181
Query: 1089 KFNLNVQAVNV 1099
KFNLNVQAVNV
Sbjct: 182 KFNLNVQAVNV 214
>TC221731 UP|Q39834 (Q39834) Clathrin heavy chain, partial (4%)
Length = 636
Score = 117 bits (293), Expect = 4e-26
Identities = 59/69 (85%), Positives = 62/69 (89%), Gaps = 1/69 (1%)
Frame = +3
Query: 1638 EVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
+VKAKEQEEKEVIAQQNMYAQLLPLALPAPPMP GMGGG+APPPPM GG+GMPPMPP
Sbjct: 3 QVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMP----GMGGGFAPPPPM-GGLGMPPMPP 167
Query: 1698 FGM-PMGGY 1705
FGM PMG Y
Sbjct: 168 FGMPPMGSY 194
Score = 30.4 bits (67), Expect = 7.0
Identities = 15/32 (46%), Positives = 17/32 (52%)
Frame = -3
Query: 1668 PMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFG 1699
PM G+ G GG PP GGG PP+P G
Sbjct: 187 PMGGIPKGGIGGIPNPPIGGGGAKPPPIPGIG 92
>TC215054 homologue to UP|Q39834 (Q39834) Clathrin heavy chain, partial (3%)
Length = 581
Score = 112 bits (281), Expect = 1e-24
Identities = 53/63 (84%), Positives = 58/63 (91%), Gaps = 2/63 (3%)
Frame = +3
Query: 1644 QEEKEVIAQQNMYAQLLPLALPAPPMPGMG--GGMGGGYAPPPPMGGGMGMPPMPPFGMP 1701
QEEK+V+AQQNMYAQLLPLALPAPPMPGMG GGMGGG+APPPPM GG+GMPPMPP+GMP
Sbjct: 3 QEEKDVVAQQNMYAQLLPLALPAPPMPGMGGPGGMGGGFAPPPPM-GGLGMPPMPPYGMP 179
Query: 1702 MGG 1704
G
Sbjct: 180 PMG 188
Score = 34.3 bits (77), Expect = 0.48
Identities = 19/37 (51%), Positives = 21/37 (56%)
Frame = -2
Query: 1668 PMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
PM GM G GG PP GGG PP+PP G P+ G
Sbjct: 187 PMGGMP*GGIGGIPSPPIGGGGAKPPPIPP-GPPIPG 80
>AW705524
Length = 370
Score = 112 bits (279), Expect = 2e-24
Identities = 66/131 (50%), Positives = 80/131 (60%), Gaps = 5/131 (3%)
Frame = +2
Query: 1579 ECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGKVDELVKHKIESQNE 1638
E +SCLFV +DLI + +EL MHNM D+AFPYL + +YT E V + QN
Sbjct: 8 EFLSSCLFVSFDLILAYIGVELTMMHNMTDYAFPYLSHILPKYTV---EFVNTN-DGQN* 175
Query: 1639 V-----KAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMP 1693
+ + KE E +VIA NMYAQLLPLA PAPPMP M GG+ PPM G+GMP
Sbjct: 176 MHLMK*RLKEHEHNDVIALHNMYAQLLPLAWPAPPMPDM-----GGFTSSPPM-CGLGMP 337
Query: 1694 PMPPFGMPMGG 1704
MPP+GMP G
Sbjct: 338 LMPPYGMPPMG 370
>AI965766
Length = 238
Score = 84.7 bits (208), Expect = 3e-16
Identities = 43/77 (55%), Positives = 56/77 (71%)
Frame = +2
Query: 8 PILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPNQPLRRPI 67
PI MRE L+LPSIGI+ Q I FTH ++D +I RET P N V I+DM+MP++ L R I
Sbjct: 8 PIAMREVLSLPSIGIHAQFINFTHGKNQADTHISGRETIPLNRVGIIDMSMPHKALTRTI 187
Query: 68 TADSALMNPNSRILALK 84
TAD A++NP+ RIL L+
Sbjct: 188TADWAIVNPHDRILDLR 238
>TC212998 similar to UP|Q8L3R8 (Q8L3R8) AT3g08530/T8G24_1, partial (4%)
Length = 568
Score = 76.6 bits (187), Expect = 8e-14
Identities = 40/52 (76%), Positives = 41/52 (77%), Gaps = 3/52 (5%)
Frame = +3
Query: 1657 AQLLPLALPAPPMPGMGGGMGGGYAPPPPMG-GGMGMPPMPPFGMPM--GGY 1705
AQLLPLALPAPPM GMGGG YAP PP+ GGMGMPPMPPFGMP GGY
Sbjct: 3 AQLLPLALPAPPMQGMGGG----YAPSPPLPMGGMGMPPMPPFGMPPMDGGY 146
>TC215051 homologue to UP|Q39834 (Q39834) Clathrin heavy chain, partial (4%)
Length = 563
Score = 71.6 bits (174), Expect = 3e-12
Identities = 34/34 (100%), Positives = 34/34 (100%)
Frame = +1
Query: 1500 DLHDNFDQIGLAQKIEKHELLEMRRVAAYIYKKA 1533
DLHDNFDQIGLAQKIEKHELLEMRRVAAYIYKKA
Sbjct: 19 DLHDNFDQIGLAQKIEKHELLEMRRVAAYIYKKA 120
Score = 48.5 bits (114), Expect = 2e-05
Identities = 20/26 (76%), Positives = 23/26 (87%)
Frame = +3
Query: 1679 GYAPPPPMGGGMGMPPMPPFGMPMGG 1704
G+APPPPMGG +GMPPMPP+GMP G
Sbjct: 117 GFAPPPPMGG-LGMPPMPPYGMPPMG 191
>BI969668 similar to SP|P49951|CLH_ Clathrin heavy chain. [Bovine] {Bos
taurus}, partial (5%)
Length = 542
Score = 44.7 bits (104), Expect = 4e-04
Identities = 31/93 (33%), Positives = 44/93 (46%), Gaps = 2/93 (2%)
Frame = -1
Query: 1593 RVDVALELAWMHNMIDFAFPYLLQFIREYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQ 1652
R DV LE AW N++DFA PY + ++E KVD+L +++EE AQ
Sbjct: 542 RPDVVLETAWRPNIMDFAMPYFIXVMKEXLAKVDKL-------DGSESLRKEEELGSEAQ 384
Query: 1653 QNMYAQ--LLPLALPAPPMPGMGGGMGGGYAPP 1683
+Y Q L+ P+ +P G GY P
Sbjct: 383 PIVYGQPHLMVXTGPSVAVPPQ-APFGYGYRAP 288
>TC204273 similar to UP|Q8W2C2 (Q8W2C2) Phosphoenolpyruvate carboxykinase
(Fragment), partial (89%)
Length = 1130
Score = 37.4 bits (85), Expect = 0.057
Identities = 30/105 (28%), Positives = 44/105 (41%), Gaps = 16/105 (15%)
Frame = +3
Query: 734 LSSSEDPDIHFKYIEAAAKTGQIKE---VERVTRESSFYDPEKTKN--------FLMEAK 782
LS ++PDI A K G + E + TRE F D T+N ++ AK
Sbjct: 147 LSKDKEPDIW-----NAIKFGTVLENVVFDEHTREVDFSDKSVTENTRAAYPIEYIPNAK 311
Query: 783 LPDARP-----LINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKV 822
+P P ++ CD FG +P ++ M +I GY V
Sbjct: 312 IPCVAPHAKNVILLACDAFGVLPPVSKLSLAQTMYHFISGYTALV 446
>AI959933 weakly similar to GP|27461187|gb type IV collagen alpha 5 chain
{Canis familiaris}, partial (2%)
Length = 380
Score = 35.8 bits (81), Expect = 0.17
Identities = 18/35 (51%), Positives = 20/35 (56%), Gaps = 2/35 (5%)
Frame = -3
Query: 1671 GMGGGMGGG--YAPPPPMGGGMGMPPMPPFGMPMG 1703
G GG GGG + PPPP GGG P P G P+G
Sbjct: 372 GCVGGKGGGPPFPPPPPPGGGGFFLPGGPPGFPLG 268
Score = 35.8 bits (81), Expect = 0.17
Identities = 19/36 (52%), Positives = 19/36 (52%), Gaps = 3/36 (8%)
Frame = +2
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGM---PPMPP 1697
PAP G GG G PPPP GGG G PP PP
Sbjct: 257 PAPFPRGKPGGPPGKKKPPPPGGGGGGKGGPPPFPP 364
Score = 33.9 bits (76), Expect = 0.63
Identities = 19/42 (45%), Positives = 20/42 (47%), Gaps = 5/42 (11%)
Frame = -3
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPM-----PPFGMP 1701
P PP PG GG G P P+G G G PP PP G P
Sbjct: 336 PPPPPPGGGGFFLPGGPPGFPLGKGAGGPPPFFGVGPPLGPP 211
Score = 31.2 bits (69), Expect = 4.1
Identities = 17/39 (43%), Positives = 17/39 (43%)
Frame = -3
Query: 1661 PLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFG 1699
P P P P GGG GGG PP PP P FG
Sbjct: 213 PRVYPFSPAPVFGGGWGGGTLFPPG-------PPFPFFG 118
>TC228704 similar to UP|Q9FJ12 (Q9FJ12) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K21P3 (Extensin like protein),
partial (42%)
Length = 1003
Score = 35.8 bits (81), Expect = 0.17
Identities = 20/44 (45%), Positives = 21/44 (47%), Gaps = 4/44 (9%)
Frame = +3
Query: 1665 PAPPMPGMGGGMGGG----YAPPPPMGGGMGMPPMPPFGMPMGG 1704
P PP P GG GGG Y+PPPP PP PP GG
Sbjct: 345 PPPPSPPSSGGGGGGAPYYYSPPPPSQYTYSSPP-PPASTGGGG 473
Score = 35.0 bits (79), Expect = 0.28
Identities = 16/32 (50%), Positives = 16/32 (50%)
Frame = +3
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMP 1696
P PP GGG GG Y PPP G PP P
Sbjct: 441 PPPPASTGGGGGGGIYYYPPPSNGNYPRPPPP 536
Score = 30.4 bits (67), Expect = 7.0
Identities = 16/33 (48%), Positives = 16/33 (48%), Gaps = 2/33 (6%)
Frame = +3
Query: 1667 PPMPGMGGGMGGG--YAPPPPMGGGMGMPPMPP 1697
PP GGG GGG Y PPP G PP P
Sbjct: 444 PPPASTGGGGGGGIYYYPPPSNGNYPRPPPPNP 542
>BG237286 similar to PIR|T10586|T105 small nuclear ribonucleoprotein-associated
protein homolog F9F13.90 - Arabidopsis thaliana, partial
(14%)
Length = 425
Score = 35.8 bits (81), Expect = 0.17
Identities = 20/40 (50%), Positives = 22/40 (55%)
Frame = +2
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
PAPP PGM APPPP G MPP P G+P+ G
Sbjct: 113 PAPPRPGMP-------APPPPRPG---MPPPPGSGVPVFG 202
Score = 30.8 bits (68), Expect = 5.3
Identities = 17/36 (47%), Positives = 18/36 (49%), Gaps = 3/36 (8%)
Frame = +2
Query: 1665 PAPPMPGMGGGMGGG---YAPPPPMGGGMGMPPMPP 1697
P PP PGM G G + PP P GM PP PP
Sbjct: 143 PPPPRPGMPPPPGSGVPVFGPPRP---GMPPPPNPP 241
>AW350216 similar to PIR|T10586|T105 small nuclear ribonucleoprotein-associated
protein homolog F9F13.90 - Arabidopsis thaliana, partial
(12%)
Length = 538
Score = 35.8 bits (81), Expect = 0.17
Identities = 20/40 (50%), Positives = 22/40 (55%)
Frame = -2
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
PAPP PGM APPPP G MPP P G+P+ G
Sbjct: 453 PAPPRPGMP-------APPPPRPG---MPPPPGSGVPVFG 364
Score = 30.8 bits (68), Expect = 5.3
Identities = 17/36 (47%), Positives = 18/36 (49%), Gaps = 3/36 (8%)
Frame = -2
Query: 1665 PAPPMPGMGGGMGGG---YAPPPPMGGGMGMPPMPP 1697
P PP PGM G G + PP P GM PP PP
Sbjct: 423 PPPPRPGMPPPPGSGVPVFGPPRP---GMPPPPNPP 325
>TC229023
Length = 1337
Score = 35.4 bits (80), Expect = 0.22
Identities = 18/36 (50%), Positives = 27/36 (75%), Gaps = 1/36 (2%)
Frame = +3
Query: 1470 VQSNNVSAVNEALNEIYVEEEDY-DRLRESIDLHDN 1504
+Q NNV+A NEA +E +E E++ D L+ESID+ +N
Sbjct: 246 LQINNVTAENEACHEHLIEMENFVDSLKESIDIAEN 353
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 65,825,335
Number of Sequences: 63676
Number of extensions: 911889
Number of successful extensions: 6249
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 5099
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5926
length of query: 1705
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1595
effective length of database: 5,635,272
effective search space: 8988258840
effective search space used: 8988258840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC142498.7


