

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.3 - phase: 0
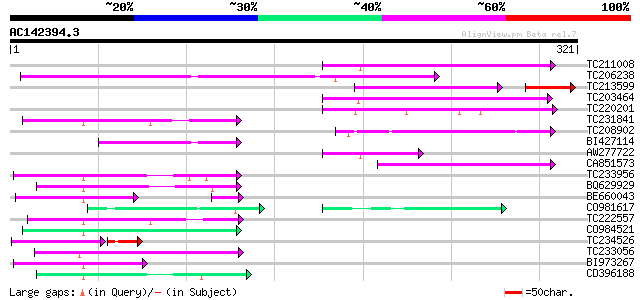
(321 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211008 100 9e-22
TC206238 weakly similar to UP|Q7PX96 (Q7PX96) AgCP12237 (Fragmen... 98 4e-21
TC213599 homologue to UP|EXBB_PASHA (P72202) Biopolymer transpor... 68 2e-17
TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leish... 81 7e-16
TC220201 similar to UP|Q9ERI5 (Q9ERI5) Apoptotic cell clearance ... 67 8e-12
TC231841 55 6e-08
TC208902 54 9e-08
BI427114 54 1e-07
AW277722 similar to SP|O65719|HS73_ Heat shock cognate 70 kDa pr... 53 2e-07
CA851573 51 8e-07
TC233956 51 8e-07
BQ629929 50 1e-06
BE660043 47 2e-06
CO981617 36 4e-06
TC222557 48 5e-06
CO984521 48 5e-06
TC234526 similar to UP|Q9XH61 (Q9XH61) Serine carboxypeptidase, ... 44 6e-06
TC233056 47 9e-06
BI973267 47 9e-06
CD396188 weakly similar to GP|6175788|gb|A ORF144 {Xestia c-nigr... 45 3e-05
>TC211008
Length = 516
Score = 100 bits (249), Expect = 9e-22
Identities = 52/137 (37%), Positives = 82/137 (58%), Gaps = 5/137 (3%)
Frame = +1
Query: 178 WKEPAIRWVKINTDGACKDG-----SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELW 232
W+ P WV +NTDG+ + S + CGGL+R S G +L GF+ LG +AELW
Sbjct: 64 WRCPXNGWVCLNTDGSVFENHRNGCSGSACGGLVRDSSGCYLGGFTVNLGNTSVTLAELW 243
Query: 233 GVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVK 292
GV+ GL+ A +G V++++DS + ++ +N +LV +I +L++ EW V+
Sbjct: 244 GVVHGLKLAWDLGCKKVKVDIDSGNALGLVRHGPVANDPAFALVSEINELVRKEWLVEFS 423
Query: 293 HSYREANRCADALANIG 309
H +RE+NR AD LA++G
Sbjct: 424 HVFRESNRAADKLAHLG 474
>TC206238 weakly similar to UP|Q7PX96 (Q7PX96) AgCP12237 (Fragment), partial
(9%)
Length = 1259
Score = 98.2 bits (243), Expect = 4e-21
Identities = 70/246 (28%), Positives = 111/246 (44%), Gaps = 9/246 (3%)
Frame = +3
Query: 7 NDAQDEIWLKLWNIRAPERIKHFMSMLYHGRLSTN-LRKHKMGIGNPMCRFCHDEIESEI 65
N ++ +L PERI + + G L TN LR+++ + C C + E+ +
Sbjct: 81 NSMLSRLFCRLSRWVGPERIINLLGNAADGALFTNPLRRYRHTSMDSSCPRCPELEETCL 260
Query: 66 HVLRDCPKATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHT 125
H LRDCPK A W V+ FF GD++ W++ NL + W F+ A
Sbjct: 261 HALRDCPKVAAFWRSVLPKKLAPKFFNGDVAVWLETNL---SFSEAAFFWPTFFGIAVEL 431
Query: 126 IWNWRNKET-HNDNYQRPLHAKNIIMIYVNDYHNAIAKFVIVTSQPRHLEEVGWKEPAI- 183
+W RN + D L +I I N Y + + PR+L + W+ P
Sbjct: 432 LWESRNDLVFYKDGTWDYLDLSDITDIVFNRYKDCMRAHASHILMPRNL--LKWRRPLPL 605
Query: 184 ---RW-VKINTDGAC-KDGSIAGCGGLIRGSEGEWLAGFSKFLGKC-DAFIAELWGVLEG 237
W +++N GA + A CGG+ R + ++ GFS LG+C E+WG+ G
Sbjct: 606 SHGHWLLRLNVSGAYDRSSDTAACGGIFRDNNDRFVLGFSVKLGECLSNDEGEIWGIYHG 785
Query: 238 LRCAKR 243
++ A+R
Sbjct: 786 MKIARR 803
>TC213599 homologue to UP|EXBB_PASHA (P72202) Biopolymer transport exbB
protein, partial (8%)
Length = 825
Score = 68.2 bits (165), Expect(2) = 2e-17
Identities = 38/84 (45%), Positives = 48/84 (56%)
Frame = +3
Query: 196 DGSIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDS 255
D +A CGGL+R G W A F LG AF+AEL G GL A G +E+N+DS
Sbjct: 339 DNDMAACGGLLRDCHGRWAASFV*KLGNTSAFVAEL*GAYLGLSLAWDKGDKNLEVNIDS 518
Query: 256 LVVVNIITSERESNASGRSLVQKI 279
LVVV I ++ +ASG SL+ I
Sbjct: 519 LVVVVSIKISKQGSASGWSLINTI 590
Score = 38.1 bits (87), Expect(2) = 2e-17
Identities = 17/28 (60%), Positives = 18/28 (63%)
Frame = +2
Query: 293 HSYREANRCADALANIGCIMGNEMMFYE 320
H YREAN CADALAN C M E + E
Sbjct: 629 HIYREANGCADALANYACAMEEEYVILE 712
>TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leishmania
major;} , partial (8%)
Length = 1386
Score = 80.9 bits (198), Expect = 7e-16
Identities = 50/134 (37%), Positives = 74/134 (54%), Gaps = 4/134 (2%)
Frame = +2
Query: 178 WKEPAIRWVKINTDGACKD--GSIAGCGGLIRGSEGEWLAGFSKFLGKCDA-FIAELWGV 234
WK+P I WVK+N DG+ S AGCGG++R + +WL GF+K L A EL +
Sbjct: 488 WKKPEIGWVKLNVDGSRDPYKSSSAGCGGVLRDASAKWLRGFAKKLNPTYAVHQTELEAI 667
Query: 235 LEGLRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIR-KLLQMEWKVKVKH 293
L GL+ A M + + DS VV+++ + + N +V+ IR K + +W+V+
Sbjct: 668 LTGLKVASEMNVKKLIVESDSDSVVSMVENGVKPNHPDYGVVELIRTKRRRFDWEVRFVS 847
Query: 294 SYREANRCADALAN 307
+ANR AD LAN
Sbjct: 848 VSNKANRVADRLAN 889
>TC220201 similar to UP|Q9ERI5 (Q9ERI5) Apoptotic cell clearance receptor
PtdSerR (Phosphatidylserine receptor), partial (5%)
Length = 1185
Score = 67.4 bits (163), Expect = 8e-12
Identities = 50/143 (34%), Positives = 75/143 (51%), Gaps = 10/143 (6%)
Frame = +1
Query: 178 WKEPAIRWVKINTDGAC--KDGSIAGCGGLIRGSEGEWLAGFSKFLGK--C-DAFIAELW 232
WK+P WVK+N DG+ ++ + AGCGG+IR G W GF + L C A EL
Sbjct: 337 WKKPESGWVKLNVDGSRIHEEPASAGCGGVIRDEWGTWCVGFDQKLDPNICRQAHYTELQ 516
Query: 233 GVLEGLRCAKRMGFTAVELNV--DSLVVVNIITSE--RESNASGRSLVQKIRKLLQ-MEW 287
+L GL+ A+ +L V DS VN++ S + + +VQ I +LL+ +W
Sbjct: 517 AILTGLKVAREDMINVEKLVVESDSEPAVNMVKSRLGYKYHRPEYKVVQDINRLLKDPKW 696
Query: 288 KVKVKHSYREANRCADALANIGC 310
++ + R+ANR A+ LA C
Sbjct: 697 LARLDYVPRKANRVANYLAKRVC 765
>TC231841
Length = 791
Score = 54.7 bits (130), Expect = 6e-08
Identities = 35/135 (25%), Positives = 56/135 (40%), Gaps = 11/135 (8%)
Frame = +3
Query: 8 DAQDEIWLKLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDEIESEI 65
+ QD ++ +LW ++ P +I F L RL T NLR+ ++ + +P C FC ES
Sbjct: 147 EQQDGVFEELWKLKLPSKITIFAWRLIRDRLPTRSNLRRKQIEVDDPRCPFCRSAEESAA 326
Query: 66 HVLRDCPKATALW---------LCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWR 116
H+ C + +W L V N R F + +++ W+
Sbjct: 327 HLFFHCSRIAPVWWESLSWVNLLGVFPNHPRQHFLQ---------HIYGVTAGMRASRWK 479
Query: 117 DFWATACHTIWNWRN 131
+W TIW RN
Sbjct: 480 WWWLALTWTIWKQRN 524
>TC208902
Length = 838
Score = 53.9 bits (128), Expect = 9e-08
Identities = 40/127 (31%), Positives = 62/127 (48%), Gaps = 2/127 (1%)
Frame = +1
Query: 185 WVKINT--DGACKDGSIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAK 242
W +IN+ DGA G +A CG ++R G + F+ LG C AE+W + G++ A
Sbjct: 223 WGRINSNCDGAVS-GGVAACGRVLRNQAGAVVV-FAHNLGMCSISQAEIWAITMGVKLAW 396
Query: 243 RMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCA 302
GFT + + D + ++ E + + LV+ + + E H REAN+ A
Sbjct: 397 DKGFTNLYVESDFKYAIEMMDGVCEFSNNCFQLVKSAKDSV*GE-TYSWCHILREANKTA 573
Query: 303 DALANIG 309
DALA G
Sbjct: 574 DALAKFG 594
>BI427114
Length = 425
Score = 53.5 bits (127), Expect = 1e-07
Identities = 26/81 (32%), Positives = 37/81 (45%)
Frame = +3
Query: 51 NPMCRFCHDEIESEIHVLRDCPKATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWN 110
N C C E+ +H LRDCPK W V+ FF GD++ W++ NL +
Sbjct: 6 NSSCARCDFLEETCLHALRDCPKVAEFWRSVIPKKLAPKFFNGDVTVWLERNL---SFSE 176
Query: 111 NNVEWRDFWATACHTIWNWRN 131
W F+ A +W +RN
Sbjct: 177 AAFFWPTFFGIAVELLWEFRN 239
>AW277722 similar to SP|O65719|HS73_ Heat shock cognate 70 kDa protein 3
(Hsc70.3). [Mouse-ear cress] {Arabidopsis thaliana},
partial (8%)
Length = 432
Score = 53.1 bits (126), Expect = 2e-07
Identities = 27/62 (43%), Positives = 36/62 (57%), Gaps = 5/62 (8%)
Frame = +1
Query: 178 WKEPAIRWVKINTDGACKDG-----SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELW 232
W+ P WV +NTDG+ + S + CGGL+R S G +L GF+ LG +AELW
Sbjct: 247 WRCPPNGWVCLNTDGSVFENHRNGCSGSACGGLVRDSSGCYLGGFTVNLGNTSVTLAELW 426
Query: 233 GV 234
GV
Sbjct: 427 GV 432
>CA851573
Length = 614
Score = 50.8 bits (120), Expect = 8e-07
Identities = 33/102 (32%), Positives = 51/102 (49%), Gaps = 1/102 (0%)
Frame = +1
Query: 209 SEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERES 268
S E L+ FSK LG C+ +AELW + G++ A + +D+ V++I +
Sbjct: 40 SIAEXLSCFSKRLGYCNVILAELWSLKYGIKLAIDLNTQFSLFEMDAREAVHMIDXIKSV 219
Query: 269 NASGRSLVQKIRKLLQME-WKVKVKHSYREANRCADALANIG 309
+S R L IR++L W+ + H REAN A+ A G
Sbjct: 220 PSSLRPLAADIRRVLWANGWQSSIVHINREANSAANWAAKFG 345
>TC233956
Length = 757
Score = 50.8 bits (120), Expect = 8e-07
Identities = 37/145 (25%), Positives = 59/145 (40%), Gaps = 16/145 (11%)
Frame = +2
Query: 3 SNFDNDAQDEIWLKLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDE 60
S D++ Q + KLW I+ P + F+ L RL T NL K ++ + + +C FCH +
Sbjct: 128 SELDDEGQHLGFKKLWEIKVPPKALSFVWRLLWDRLPTKDNLIKRQIQVEDDLCPFCHSQ 307
Query: 61 IESEIHVLRDCPKATALWLCVVDNAARTSFFEGDLSSWIK---------FNLFTNVYWN- 110
E+ H+ C K LW + SW+K + F Y +
Sbjct: 308 SETASHLFFTCGKIMPLW--------------WEFLSWVKEDKVFHCRPMDNFLQHYSSA 445
Query: 111 ----NNVEWRDFWATACHTIWNWRN 131
+N +W ++IW RN
Sbjct: 446 ASKVSNTRRTMWWIAVTNSIWRLRN 520
>BQ629929
Length = 437
Score = 50.4 bits (119), Expect = 1e-06
Identities = 34/132 (25%), Positives = 54/132 (40%), Gaps = 16/132 (12%)
Frame = +3
Query: 16 KLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDEIESEIHVLRDCPK 73
+LW I+ P R F L RL T NL + ++ I N +C FC ++ES H+ K
Sbjct: 15 QLWEIKVPPRALSFAWRLLWDRLPTKENLIRRQVVIENDLCPFCQSQVESASHLFFTYKK 194
Query: 74 ATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNV--------------EWRDFW 119
+W + +SW+K + + +N +W+ +W
Sbjct: 195 VMPMW*--------------EFNSWVKEDTVMHCRPGDNFLQHFSLAGSRDSNRKWKIWW 332
Query: 120 ATACHTIWNWRN 131
A +IWN RN
Sbjct: 333 IAATTSIWNLRN 368
>BE660043
Length = 714
Score = 47.0 bits (110), Expect(2) = 2e-06
Identities = 23/72 (31%), Positives = 37/72 (50%), Gaps = 2/72 (2%)
Frame = -2
Query: 4 NFDNDAQDEIWLKLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDEI 61
N + D+I+ LW ++ P + F L RL T N+RK ++ I + +C FC +
Sbjct: 554 NSEEGNMDDIFKDLWKLKIPAKASIFAGRLIRDRLPTKSNMRKRQIDINDSLCPFCSIKE 375
Query: 62 ESEIHVLRDCPK 73
E+ H+ DC K
Sbjct: 374 ETASHLFFDCSK 339
Score = 22.3 bits (46), Expect(2) = 2e-06
Identities = 7/18 (38%), Positives = 10/18 (54%)
Frame = -1
Query: 115 WRDFWATACHTIWNWRNK 132
W+ +W +IW RNK
Sbjct: 216 WKCWWVALNWSIWQHRNK 163
>CO981617
Length = 837
Score = 35.8 bits (81), Expect(2) = 4e-06
Identities = 26/104 (25%), Positives = 40/104 (38%)
Frame = -1
Query: 178 WKEPAIRWVKINTDGACKDGSIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEG 237
W P K+N DG I + W+A C A+LW + G
Sbjct: 351 WSPPCPGVSKVNCDGVV----------FIHEAIATWMAS-------CSILEAKLWSIFYG 223
Query: 238 LRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIRK 281
+R A+ G + + DS+ VN + N LVQ+I++
Sbjct: 222 MRLAEDCGVQHIIMESDSVEAVNHLMEHSYDNHPLFHLVQEIKR 91
Score = 32.0 bits (71), Expect(2) = 4e-06
Identities = 26/110 (23%), Positives = 42/110 (37%), Gaps = 10/110 (9%)
Frame = -3
Query: 45 HKMGIGNPMCRFCHDEIESEIHVLRDCPKATALWLCVVDNAARTSFFEGDLSSWIKFNLF 104
+ +G P C+ +IE+ RDC + +W + FF GDL++W+ N
Sbjct: 748 YDLGKXLP*CK----KIETLTRCFRDCYRVRKVWDSLWSGRKPPIFFHGDLNTWMTANPK 581
Query: 105 TNVYWNNNVEWRDFWATACHTI----------WNWRNKETHNDNYQRPLH 144
N Y N W + I W ++E +N Q +H
Sbjct: 580 GN-YDNRISNWSQCFGVTLDRI**RINDYVFGQKWTSEEGIINNVQTMMH 434
>TC222557
Length = 1002
Score = 48.1 bits (113), Expect = 5e-06
Identities = 33/133 (24%), Positives = 55/133 (40%), Gaps = 11/133 (8%)
Frame = +1
Query: 11 DEIWLKLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDEIESEIHVL 68
DE LW ++ P + F L R+ T NL + ++ + N MC FC+ + E H+
Sbjct: 421 DEALEDLWQLKIPLKATTFAWRLIKERIPTKGNLWRRRVQLNNLMCPFCNRQEEEASHLF 600
Query: 69 RDCPKATALW---------LCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFW 119
+CP+ LW + V R ++ + L S K + W+ +W
Sbjct: 601 FNCPRILPLWWESLAWTKTVGVFSPIPRQNYMQHTLISKGK---------HLQARWKCWW 753
Query: 120 ATACHTIWNWRNK 132
+IW RN+
Sbjct: 754 VALTWSIWRHRNR 792
>CO984521
Length = 716
Score = 48.1 bits (113), Expect = 5e-06
Identities = 30/126 (23%), Positives = 50/126 (38%), Gaps = 2/126 (1%)
Frame = -2
Query: 8 DAQDEIWLKLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDEIESEI 65
+ QD + +LW +R P ++ F L +L T NLRK ++ + +C C E+
Sbjct: 562 EEQDGKFKELWKLRVPLKVAIFAWRLIQDKLPTKANLRKKRVELQEYLCPLCRSVEETAS 383
Query: 66 HVLRDCPKATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHT 125
H+ C K + LW F ++F W+ +W ++
Sbjct: 382 HLFFHCSKVSPLWWESQSWVNMMGVFPYQPDQHFSQHIFGASVGLQGKRWQWWWFALTYS 203
Query: 126 IWNWRN 131
IW RN
Sbjct: 202 IWKHRN 185
>TC234526 similar to UP|Q9XH61 (Q9XH61) Serine carboxypeptidase, partial
(11%)
Length = 603
Score = 43.5 bits (101), Expect(2) = 6e-06
Identities = 20/53 (37%), Positives = 29/53 (53%)
Frame = +1
Query: 2 LSNFDNDAQDEIWLKLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIGNPMC 54
++ F + E W K+W I AP++I+ F+ L H RL+T R GIG C
Sbjct: 73 INQFHTLNESENWKKIWKINAPQKIRVFIWQLVHKRLTTKKRVVSWGIGTLSC 231
Score = 23.9 bits (50), Expect(2) = 6e-06
Identities = 11/20 (55%), Positives = 15/20 (75%)
Frame = +3
Query: 56 FCHDEIESEIHVLRDCPKAT 75
F H E E+ +HV+RDCP A+
Sbjct: 237 FRHFE-ETILHVMRDCPLAS 293
>TC233056
Length = 680
Score = 47.4 bits (111), Expect = 9e-06
Identities = 28/120 (23%), Positives = 51/120 (42%), Gaps = 2/120 (1%)
Frame = -1
Query: 15 LKLWNIRAPERIKHFMSMLYHGRL--STNLRKHKMGIGNPMCRFCHDEIESEIHVLRDCP 72
++LW ++ P + F L RL TNLR+ ++ + + +C FC ++ E H+ +
Sbjct: 665 VELWKLKIPAKSAVFAWRLIRDRLPTKTNLRRRQVMLTDSLCPFCRNKEEEATHLFFNYS 486
Query: 73 KATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNK 132
LW + T+ + + N + + W+ +W TIW RNK
Sbjct: 485 NILPLWWESLS*VNLTTALPQNPRDHYLQHGIGNDHGKKAIRWKCWWIALTRTIWQHRNK 306
>BI973267
Length = 406
Score = 47.4 bits (111), Expect = 9e-06
Identities = 26/78 (33%), Positives = 40/78 (50%), Gaps = 2/78 (2%)
Frame = +1
Query: 3 SNFDNDAQDEIWLKLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDE 60
S D+ Q E + +L NI+ P + F L RL T NL K ++ + N +C FCH++
Sbjct: 130 SELDDQGQYEGFQQLCNIKIPPKALSFAWRLL*DRLPTKDNLVKRQIQVENDLCPFCHNQ 309
Query: 61 IESEIHVLRDCPKATALW 78
E+ H+ C K +W
Sbjct: 310 PETASHLFFKCDKIMPMW 363
>CD396188 weakly similar to GP|6175788|gb|A ORF144 {Xestia c-nigrum
granulovirus}, partial (5%)
Length = 701
Score = 45.4 bits (106), Expect = 3e-05
Identities = 32/138 (23%), Positives = 55/138 (39%), Gaps = 16/138 (11%)
Frame = -1
Query: 16 KLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDEIESEIHVLRDCPK 73
+LW I+ P F L RL + NL + ++ + N +C FC ++ES H+ C K
Sbjct: 629 QLWEIKIPPTALSFAWRLLWDRLPSKENLIRRQIVLQNDLCPFCQSQVESASHLFFTCHK 450
Query: 74 ATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNV--------------YWNNNVEWRDFW 119
LW + ++W++ + + N+N + +W
Sbjct: 449 VMPLW--------------WEFNTWVREDRVLHSKPMDNFLQHCSLAGSRNSNRRRKIWW 312
Query: 120 ATACHTIWNWRNKETHND 137
A +IWN RN N+
Sbjct: 311 IAATRSIWNLRNDMIFNN 258
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.137 0.450
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,094,011
Number of Sequences: 63676
Number of extensions: 325107
Number of successful extensions: 1682
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 1636
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1660
length of query: 321
length of database: 12,639,632
effective HSP length: 97
effective length of query: 224
effective length of database: 6,463,060
effective search space: 1447725440
effective search space used: 1447725440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC142394.3


