

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.1 - phase: 0 /pseudo
(426 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
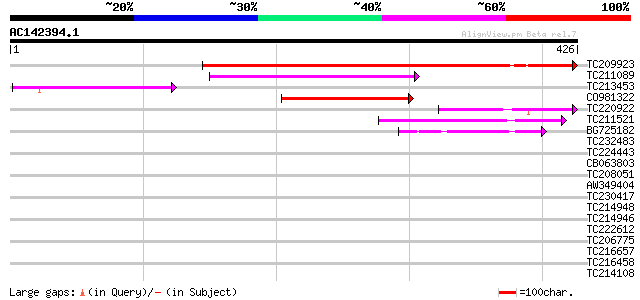
Score E
Sequences producing significant alignments: (bits) Value
TC209923 weakly similar to UP|Q84UV0 (Q84UV0) Linalool synthase,... 418 e-117
TC211089 weakly similar to UP|O23517 (O23517) LIMONENE cyclase l... 83 3e-16
TC213453 weakly similar to UP|O64961 (O64961) Epidermal germacre... 74 1e-13
CO981322 73 3e-13
TC220922 weakly similar to UP|Q940E7 (Q940E7) D-limonene synthas... 54 1e-07
TC211521 weakly similar to UP|Q9FRX5 (Q9FRX5) Ent-kaurene syntha... 48 7e-06
BG725182 weakly similar to GP|2465021|emb| sesquiterpene cyclase... 42 4e-04
TC232483 UP|O82687 (O82687) Tyrosine phosphatase 1, complete 32 0.54
TC224443 30 2.7
CB063803 29 3.5
TC208051 weakly similar to UP|Q8S341 (Q8S341) Purple acid phosph... 29 3.5
AW349404 29 3.5
TC230417 similar to UP|Q9C5V6 (Q9C5V6) Topoisomerase 6 subunit B... 29 3.5
TC214948 weakly similar to UP|Q9ZVV1 (Q9ZVV1) T5A14.4 protein, p... 29 4.6
TC214946 homologue to UP|Q6TEC7 (Q6TEC7) Defender against progra... 29 4.6
TC222612 28 6.0
TC206775 similar to UP|Q7XXP9 (Q7XXP9) RNA helicase (Fragment), ... 28 6.0
TC216657 similar to UP|MHK_ARATH (P43294) Serine/threonine-prote... 28 6.0
TC216458 similar to UP|Q9WUG9 (Q9WUG9) ARL-6 interacting protein... 28 7.9
TC214108 similar to UP|RK29_ARATH (Q9FJP3) 50S ribosomal protein... 28 7.9
>TC209923 weakly similar to UP|Q84UV0 (Q84UV0) Linalool synthase, partial
(17%)
Length = 1215
Score = 418 bits (1074), Expect = e-117
Identities = 199/282 (70%), Positives = 240/282 (84%), Gaps = 1/282 (0%)
Frame = +1
Query: 146 AIYVSNTLQYPIHYGLSRFMDK-SIFINDLKAKNKWICLDELAKMNSSIVKFMNKNESVE 204
A+YV+ TLQ+P+HY LSRF D SI +ND K K +W CL+ELA++NSSIV+F+N+NE +
Sbjct: 1 AMYVAKTLQHPLHYDLSRFRDDTSILLNDFKTKREWECLEELAEINSSIVRFVNQNEITQ 180
Query: 205 VFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLVYIIDDIFDV 264
V+KWW+DLGL E+KFA Y PLKWYMWPMACFTDP FS +R+ELTKPISLVYIIDDIFDV
Sbjct: 181 VYKWWKDLGLNNEVKFARYQPLKWYMWPMACFTDPRFSEQRIELTKPISLVYIIDDIFDV 360
Query: 265 HGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLK 324
+GTLDQLTLFT+A+ RWE+ E LP+FMK+ L LY++TN+FAE +YKKHGFNPI+TLK
Sbjct: 361 YGTLDQLTLFTDAIKRWELASTEQLPDFMKMCLRVLYEITNDFAEKIYKKHGFNPIETLK 540
Query: 325 ISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETL 384
SWVRLLNAFL+EAHWLNSG LP + EYLNNGIVSTGVHVVL+H+FFLMD++ I E +
Sbjct: 541 RSWVRLLNAFLEEAHWLNSGHLPRSGEYLNNGIVSTGVHVVLVHSFFLMDYS--INNEIV 714
Query: 385 SILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+I+D P II+SVAKILRLSDDLEG KS DQNGLDGSY++C
Sbjct: 715 AIVD-NVPQIIHSVAKILRLSDDLEGAKSEDQNGLDGSYIDC 837
>TC211089 weakly similar to UP|O23517 (O23517) LIMONENE cyclase like protein,
partial (9%)
Length = 626
Score = 82.8 bits (203), Expect = 3e-16
Identities = 50/158 (31%), Positives = 81/158 (50%)
Frame = +2
Query: 151 NTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWE 210
+ L+ P H+ + F K + KN L EL K+N ++++ + E ++ +WWE
Sbjct: 107 HALELPSHWRVQWFEVKWHVEQYKQQKNVDPILLELTKLNFNMIQAKLQIEVKDLSRWWE 286
Query: 211 DLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQ 270
+LG+ KE+ FA ++ +M +P + R LTK I V I DD+ D+H + ++
Sbjct: 287 NLGIKKELSFARNRLVESFMCAAGVAFEPKYKAVRKWLTKVIIFVLINDDVDDIHASFEE 466
Query: 271 LTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFA 308
L T A RW+ E LP +MK + +L VTN A
Sbjct: 467 LKPITLAFERWDDKELEELPQYMKSCVHALKDVTNEIA 580
>TC213453 weakly similar to UP|O64961 (O64961) Epidermal germacrene C
synthase (Sesquiterpene synthase 1), partial (13%)
Length = 432
Score = 73.9 bits (180), Expect = 1e-13
Identities = 44/125 (35%), Positives = 71/125 (56%), Gaps = 2/125 (1%)
Frame = +3
Query: 3 KQVHKKLVTSEDPTENFHL--IDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSH 60
+ V K +V+ D +F L ID IQRLG+ ++F EI AL + + I + + +
Sbjct: 45 EDVRKMIVSPIDNNFSFKLNFIDSIQRLGVSYHFEHEIDRALHQIYDISTKDNNIISHDN 224
Query: 61 ELYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGED 120
+L+ VAL FRLLRQ GY + + +F K + D++G+++LYEA+QL GE+
Sbjct: 225 DLHHVALLFRLLRQHGYRITSGVFLKFKDQTGKFNERLANDIQGMLSLYEAAQLRFHGEE 404
Query: 121 GLNDL 125
L ++
Sbjct: 405 ILEEV 419
>CO981322
Length = 691
Score = 72.8 bits (177), Expect = 3e-13
Identities = 33/100 (33%), Positives = 61/100 (61%), Gaps = 1/100 (1%)
Frame = -2
Query: 205 VFKWW-EDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLVYIIDDIFD 263
VF+WW +DL ++ + F ++ W + + +P +S R + K I++ IIDD++D
Sbjct: 327 VFRWWVKDLNVSTKFPFVRDRIVECCFWILGVYIEPQYSLARRIMMKVIAISSIIDDVYD 148
Query: 264 VHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKV 303
+GT+D+L +FT+A+ RW++ +LP +MK+ S+ V
Sbjct: 147 SYGTIDELEIFTDAIERWDICSLVDLPEYMKLCYSAXXDV 28
>TC220922 weakly similar to UP|Q940E7 (Q940E7) D-limonene synthase, partial
(13%)
Length = 608
Score = 54.3 bits (129), Expect = 1e-07
Identities = 31/106 (29%), Positives = 55/106 (51%), Gaps = 2/106 (1%)
Frame = +1
Query: 323 LKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKE 382
LK +W+ A EA W N G +P+ EEYL+N +S+ V+L+ ++F +
Sbjct: 16 LKKAWIDFCKALYVEAKWYNKGYIPSLEEYLSNAWISSSGPVILLLSYF------ATMNQ 177
Query: 383 TLSILD--EEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+ I D + +++Y+V+ I+RL +DL + + G S + C
Sbjct: 178 AMDIDDFLHTYEDLVYNVSLIIRLCNDLGTTAAEREKGDVASSILC 315
>TC211521 weakly similar to UP|Q9FRX5 (Q9FRX5) Ent-kaurene synthase, partial
(24%)
Length = 980
Score = 48.1 bits (113), Expect = 7e-06
Identities = 30/143 (20%), Positives = 64/143 (43%), Gaps = 2/143 (1%)
Frame = +2
Query: 278 VNRWEMD-GAENLPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLK 336
V +W++D +K+ S+++ E K+ G N + + W+ L+ + +
Sbjct: 2 VEKWDVDINTVCCSETVKIIFSAIHSTVCEIGEKSVKQQGRNVKNNVIKIWLNLVQSMFR 181
Query: 337 EAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIY 396
EA WL + +PT +Y+ N +S + +++ A +L+ K + + + N +Y
Sbjct: 182 EAEWLRTKTVPTIGDYMENAYISFALGPIVLPALYLVG-----PKLSDEVTENHELNYLY 346
Query: 397 SVAKIL-RLSDDLEGVKSGDQNG 418
+ RL +D+ K + G
Sbjct: 347 KLMSTCGRLLNDIHSFKRESEEG 415
>BG725182 weakly similar to GP|2465021|emb| sesquiterpene cyclase {Fragaria
vesca}, partial (11%)
Length = 308
Score = 42.4 bits (98), Expect = 4e-04
Identities = 33/111 (29%), Positives = 52/111 (46%)
Frame = +3
Query: 293 MKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEY 352
MK+ S+L V E +K G N + +K RL+ A++ EA W + PT EEY
Sbjct: 3 MKLCYSALLDVFEE-TEQEMRKQGKNTLRQIK----RLVQAYITEARWFHCNHTPTMEEY 167
Query: 353 LNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILR 403
+ +S G ++ I +F M+ T E + I P I+ + + I R
Sbjct: 168 MQVATMSCGFAMLTIVSFLGMED----TTEEVLIWATSDPKIVAAASIISR 308
>TC232483 UP|O82687 (O82687) Tyrosine phosphatase 1, complete
Length = 1412
Score = 32.0 bits (71), Expect = 0.54
Identities = 19/59 (32%), Positives = 30/59 (50%)
Frame = -3
Query: 342 NSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAK 400
N I + L+ I+ST H+V+IH D+ + + ILD+ FP I+ S+ K
Sbjct: 684 NRNITNFSRPILSLKIISTFCHLVIIHQSSKHDNCRAV------ILDDHFPEILVSMLK 526
>TC224443
Length = 670
Score = 29.6 bits (65), Expect = 2.7
Identities = 13/37 (35%), Positives = 20/37 (53%)
Frame = -1
Query: 26 QRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
Q G +HY++ K +L QF+ + +D VS H L
Sbjct: 160 QGFGYQHYYMRRAKASLSNQFMKSNKINLDLVSFHLL 50
>CB063803
Length = 411
Score = 29.3 bits (64), Expect = 3.5
Identities = 12/36 (33%), Positives = 20/36 (55%)
Frame = +2
Query: 155 YPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMN 190
YP+ +S F KSI+ + + +N W D+L +N
Sbjct: 110 YPVSLPISLFQSKSIYFSSMMKQNAWT*YDKL*VLN 217
>TC208051 weakly similar to UP|Q8S341 (Q8S341) Purple acid phosphatase,
partial (84%)
Length = 1249
Score = 29.3 bits (64), Expect = 3.5
Identities = 21/95 (22%), Positives = 42/95 (44%), Gaps = 11/95 (11%)
Frame = +3
Query: 197 MNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMW-----------PMACFTDPCFSNER 245
+ KN ++ ++ W ++G KE ++P++W +W + FT
Sbjct: 135 LQKNNNLLIY-WSLEIGEEKEPTINLWSPIRWELWGRN*I*IL*SPQVTIFTKMD*KGLM 311
Query: 246 VELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNR 280
++ + +S + HGTL +T+ TEA+ R
Sbjct: 312 IQHSTSLSSTCTLPPACKRHGTLFWVTMTTEAMLR 416
>AW349404
Length = 531
Score = 29.3 bits (64), Expect = 3.5
Identities = 20/67 (29%), Positives = 35/67 (51%), Gaps = 1/67 (1%)
Frame = -2
Query: 48 ILSSNPIDFVSSHELYEV-ALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLI 106
+L+ + +D + S L+E + A RLL+QGG Y+N D + R++ + V G +
Sbjct: 485 VLNLSLVDLLVSSGLFESKSAARRLLKQGGLYLNNNRVD-----NEGKRIEAADIVDGKV 321
Query: 107 ALYEASQ 113
L A +
Sbjct: 320 LLLSAGK 300
>TC230417 similar to UP|Q9C5V6 (Q9C5V6) Topoisomerase 6 subunit B, partial
(34%)
Length = 937
Score = 29.3 bits (64), Expect = 3.5
Identities = 22/91 (24%), Positives = 41/91 (44%)
Frame = +1
Query: 93 SLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAWLPRHQDHIQAIYVSNT 152
S +++YG+D L++ + ++ E CE L H++ +
Sbjct: 496 SKKIRYGDDDAELLSKVSENLITKE----------TLCEKLSK-------HVEQVDYEMA 624
Query: 153 LQYPIHYGLSRFMDKSIFINDLKAKNKWICL 183
L+Y G+S ++I+I L+A+NK I L
Sbjct: 625 LEYATQSGVSEEPRETIYIQSLEAENKMIDL 717
>TC214948 weakly similar to UP|Q9ZVV1 (Q9ZVV1) T5A14.4 protein, partial (58%)
Length = 1024
Score = 28.9 bits (63), Expect = 4.6
Identities = 16/46 (34%), Positives = 29/46 (62%)
Frame = +1
Query: 150 SNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVK 195
S ++QYP + L RF+ +S N+ + N IC DE+ ++ SS+++
Sbjct: 874 SRSIQYPSYRDLQRFLAESTAGNE--STNSTIC-DEVCQLKSSLLE 1002
>TC214946 homologue to UP|Q6TEC7 (Q6TEC7) Defender against programmed cell
death, partial (12%)
Length = 1085
Score = 28.9 bits (63), Expect = 4.6
Identities = 16/46 (34%), Positives = 29/46 (62%)
Frame = +3
Query: 150 SNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVK 195
S ++QYP + L RF+ +S N+ + N IC DE+ ++ SS+++
Sbjct: 177 SRSIQYPSYRDLQRFLAESTAGNE--STNSTIC-DEVCQLKSSLLE 305
>TC222612
Length = 470
Score = 28.5 bits (62), Expect = 6.0
Identities = 15/62 (24%), Positives = 28/62 (44%)
Frame = -2
Query: 255 VYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKK 314
+Y I + + LD+ T+ + + GAE L +L L + NNF+ + ++
Sbjct: 385 IYKIQQLLRAYSFLDEFTIKDRNMEESSISGAEILLRSRSSTLLGLINLNNNFSNIRARE 206
Query: 315 HG 316
G
Sbjct: 205 QG 200
>TC206775 similar to UP|Q7XXP9 (Q7XXP9) RNA helicase (Fragment), partial
(36%)
Length = 445
Score = 28.5 bits (62), Expect = 6.0
Identities = 17/63 (26%), Positives = 32/63 (49%)
Frame = +1
Query: 104 GLIALYEASQLSIEGEDGLNDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSR 163
G+ L E S + ++ D + D+G+ +++++ L + Q + S T P+HY
Sbjct: 19 GICCLKEVSFVVLDEADRMLDMGFE--QIVRSILGQTCSDRQMVMFSATWPLPVHYLAQE 192
Query: 164 FMD 166
FMD
Sbjct: 193 FMD 201
>TC216657 similar to UP|MHK_ARATH (P43294) Serine/threonine-protein kinase
MHK , partial (41%)
Length = 1393
Score = 28.5 bits (62), Expect = 6.0
Identities = 27/118 (22%), Positives = 43/118 (35%), Gaps = 6/118 (5%)
Frame = +2
Query: 160 GLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMK 219
GLS K F DLK +N + D L D GLA+E+
Sbjct: 20 GLSHMHKKGFFHRDLKPENMLVTNDVLK---------------------IADFGLAREVS 136
Query: 220 ----FAGYNPLKWYMWPMACFTDPCFSN--ERVELTKPISLVYIIDDIFDVHGTLDQL 271
+ Y +WY P PC++ + + ++ ++ + IF +DQL
Sbjct: 137 SMPPYTQYVSTRWYRAPEVLLRAPCYTPAVDMWAVGAILAELFTLTPIFPGESEIDQL 310
>TC216458 similar to UP|Q9WUG9 (Q9WUG9) ARL-6 interacting protein-5
(Fragment), partial (14%)
Length = 1207
Score = 28.1 bits (61), Expect = 7.9
Identities = 10/37 (27%), Positives = 18/37 (48%)
Frame = +2
Query: 205 VFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCF 241
++ W LG+ ++ F + PL + W CF C+
Sbjct: 569 LYMWSATLGVCLDIFFCKFLPLVCFCWSPDCFMGSCY 679
>TC214108 similar to UP|RK29_ARATH (Q9FJP3) 50S ribosomal protein L29,
chloroplast precursor, partial (68%)
Length = 1202
Score = 28.1 bits (61), Expect = 7.9
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Frame = -3
Query: 280 RWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPI-DTLKISWVRLL-NAFLKE 337
RW + A LP F+K+S L NF+ ++ KH NP+ + + + L+ + FLK
Sbjct: 501 RWSYNNAF-LPFFIKLSGQPLIDTLLNFSFPLHSKHASNPLAHATEFAGLELVASRFLKA 325
Query: 338 AH 339
H
Sbjct: 324 KH 319
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,677,666
Number of Sequences: 63676
Number of extensions: 273818
Number of successful extensions: 1216
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1210
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1213
length of query: 426
length of database: 12,639,632
effective HSP length: 100
effective length of query: 326
effective length of database: 6,272,032
effective search space: 2044682432
effective search space used: 2044682432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142394.1


