

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142224.5 - phase: 0 /pseudo
(625 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
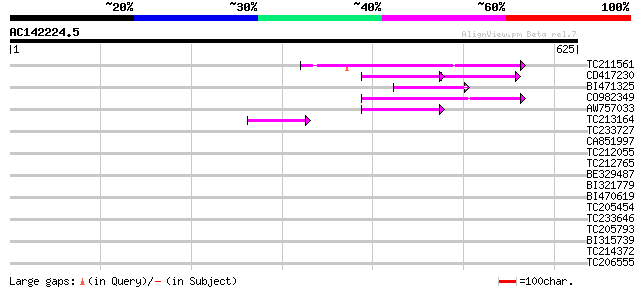
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 79 8e-15
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 56 3e-14
BI471325 66 4e-11
CO982349 65 7e-11
AW757033 57 2e-08
TC213164 50 4e-06
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 39 0.005
CA851997 37 0.020
TC212055 37 0.020
TC212765 36 0.045
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 35 0.076
BI321779 35 0.13
BI470619 33 0.50
TC205454 similar to UP|LON1_ARATH (O64948) Lon protease homolog ... 30 2.5
TC233646 30 3.2
TC205793 homologue to UP|Q9FLK9 (Q9FLK9) Leucine-rich repeat dis... 29 5.5
BI315739 28 9.4
TC214372 28 9.4
TC206555 similar to UP|U315_ARATH (Q8LFJ5) UPF0315 protein At1g2... 28 9.4
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 78.6 bits (192), Expect = 8e-15
Identities = 65/262 (24%), Positives = 109/262 (40%), Gaps = 14/262 (5%)
Frame = +1
Query: 321 LENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK--------KW- 371
L +AL EV+ K + + + K D KA+D + W++L +M + KW
Sbjct: 1 LHSALIANEVIDEAK---RSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWI 171
Query: 372 -----DSQMFGLTGSCNV*PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHG 426
+ + L P G+RQG P +P+L+ I +EGLN ++ L
Sbjct: 172 EECVKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKP 351
Query: 427 TRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRN 486
+ I+ L +ADD+ F +A +K IL ++E S IN+ KS
Sbjct: 352 YLVGANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVFGV 531
Query: 487 TDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSR 546
TD + L + YLG+P + ++ I + + + W +S
Sbjct: 532 TDQ-WKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISF 708
Query: 547 AGKEVLLKSIAQYIPTYSWAIF 568
G+ L+KS+ IP Y ++ F
Sbjct: 709 GGRVTLIKSVLTSIPIYFFSFF 774
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 56.2 bits (134), Expect(2) = 3e-14
Identities = 29/91 (31%), Positives = 48/91 (51%)
Frame = -3
Query: 389 PHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLF 448
P+ G+RQ P +PYL+++C E L+ I N L ++ RG I+ L F DD LF
Sbjct: 608 PNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL*RSIQVNRGGLLISHLAFVDDLILF 429
Query: 449 YKAYVSEDTILKNILYTYEAASGQAINYRKS 479
+A + + ++ L + +SG +N K+
Sbjct: 428 VEANMDQVDVINLTLELFSDSSGAKVNVDKT 336
Score = 40.4 bits (93), Expect(2) = 3e-14
Identities = 23/87 (26%), Positives = 42/87 (47%)
Frame = -2
Query: 477 RKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNI 536
+ +FS+N + +I+ LG+ + GKYLG+P + F FI +++ K
Sbjct: 342 QNQKFSFSKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRF 163
Query: 537 QNWSAWSLSRAGKEVLLKSIAQYIPTY 563
W + LS G+ L K + +P++
Sbjct: 162 SRWKSNLLSMVGRLTLTK*VV*ALPSH 82
>BI471325
Length = 421
Score = 66.2 bits (160), Expect = 4e-11
Identities = 37/83 (44%), Positives = 49/83 (58%)
Frame = +2
Query: 424 IHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAF 483
IH ++ RG ++ LL D FLF + E +LKNIL T+EA+SG +N RKS I F
Sbjct: 8 IHRVKVHRGITILSHLLSVDFYFLFCRTIDKEADVLKNILTTFEASSGLVVNLRKSEICF 187
Query: 484 SRNTDSIGHHNITHLLGVVESMW 506
SRNT I +I LG+ S+W
Sbjct: 188 SRNTSQIARDSIFFHLGM--SIW 250
>CO982349
Length = 795
Score = 65.5 bits (158), Expect = 7e-11
Identities = 46/180 (25%), Positives = 79/180 (43%)
Frame = +3
Query: 389 PHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLF 448
P G+RQG P +P L+ I +EGL ++ + + + + ++ L +ADD+ F
Sbjct: 141 PQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRFNSFLVGKNKEPVSILQYADDTIFF 320
Query: 449 YKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHG 508
+A + ++K IL +E ASG IN+ +S D L + SM
Sbjct: 321 EEATMENMKVIKTILRCFELASGLKINFAESRFGAIWKPDHWCKEAAEFLNCSMLSM-PF 497
Query: 509 KYLGLPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
YLG+P + I I + + W +S G+ L+ +I +P Y ++ F
Sbjct: 498 SYLGIPIEANPRCREI*DPIIRKCETKLARWKQRHISLGGRVTLINAILTALPIYFFSFF 677
>AW757033
Length = 441
Score = 57.0 bits (136), Expect = 2e-08
Identities = 30/91 (32%), Positives = 48/91 (51%)
Frame = -2
Query: 389 PHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLF 448
PH G+RQG P +P L+ I +EGL ++ + + ++ L +ADD+ F
Sbjct: 425 PHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPVSILQYADDTIFF 246
Query: 449 YKAYVSEDTILKNILYTYEAASGQAINYRKS 479
+A + ++K IL +E ASG IN+ KS
Sbjct: 245 GEATMENVRVIKTILRGFELASGLKINFAKS 153
>TC213164
Length = 446
Score = 49.7 bits (117), Expect = 4e-06
Identities = 25/69 (36%), Positives = 40/69 (57%)
Frame = +3
Query: 263 IALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFILE 322
+ALI K+ P S+ P+SL IYKI++K+L+ R K++L I + Q F+ R L
Sbjct: 213 LALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEGRHTLH 392
Query: 323 NALTYFEVL 331
N + E++
Sbjct: 393 NVVIANEIM 419
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 39.3 bits (90), Expect = 0.005
Identities = 32/90 (35%), Positives = 44/90 (48%), Gaps = 14/90 (15%)
Frame = -3
Query: 329 EVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEKKWDSQMF-----------G 377
EV++ + K G GN+ALK D+ KAFD + +L +V+ K S F
Sbjct: 807 EVINMLDKKVFG--GNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIRVILLSAK 634
Query: 378 LTGSCNV*PI---DPHCGIRQGGPFSPYLY 404
L+ S N P+ G+RQG P SP LY
Sbjct: 633 LSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
>CA851997
Length = 636
Score = 37.4 bits (85), Expect = 0.020
Identities = 25/59 (42%), Positives = 36/59 (60%), Gaps = 2/59 (3%)
Frame = +1
Query: 249 VSEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKIL--SKVLANRLKQILHK 305
++EG+FP ++ DT IA+I +I + LCN +KIL S+VLA RLK +L K
Sbjct: 397 LAEGAFPSSVNDTTIAIILEI**S*RYERS*TNLLCNGGFKILFLSEVLAKRLKNVLDK 573
>TC212055
Length = 776
Score = 37.4 bits (85), Expect = 0.020
Identities = 21/78 (26%), Positives = 39/78 (49%)
Frame = +1
Query: 403 LYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNI 462
L+ I +EGL ++ + L + + + L +ADD+ +A + +K++
Sbjct: 1 LFNIVAEGLTGLMREALDKSLYSSLMVGKNKIPVNILQYADDTIFLGEATMQNVMTIKSM 180
Query: 463 LYTYEAASGQAINYRKSS 480
L +E ASG I++ KSS
Sbjct: 181 LRVFELASGLKIHFAKSS 234
>TC212765
Length = 637
Score = 36.2 bits (82), Expect = 0.045
Identities = 35/132 (26%), Positives = 55/132 (41%)
Frame = +3
Query: 401 PYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILK 460
PYL+++C E L I + + + + + L+ DD FLF A +
Sbjct: 15 PYLFVLCMERLALCIHDLTSQ*IWRPILVSDDNRLVPYLMLVDDIFLFINAIGD*AYLAS 194
Query: 461 NILYTYEAASGQAINYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERD 520
L + G +N KSS+ S + I++ L +V + GKYL L + R
Sbjct: 195 LTL*NFFQTFGLKLNLDKSSMYCS*RMAPVLRDLISNTLNIVHAPSLGKYLCLKFIHGRS 374
Query: 521 KKSIFSFIKERI 532
K F + ERI
Sbjct: 375 KIVDFLLV-ERI 407
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 35.4 bits (80), Expect = 0.076
Identities = 16/45 (35%), Positives = 27/45 (59%)
Frame = -2
Query: 250 SEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKV 294
+ G FP + IALI K+ P+++ P+SL +YKI++K+
Sbjct: 246 ANGIFPKGGNASFIALIPKVKHPQALNDFRPISLIGCVYKIVAKI 112
>BI321779
Length = 421
Score = 34.7 bits (78), Expect = 0.13
Identities = 16/31 (51%), Positives = 20/31 (63%)
Frame = +2
Query: 263 IALIAKIDRPKSMKHLCPVSLCNVIYKILSK 293
IALIAK D P + CP+SL +YKI+ K
Sbjct: 278 IALIAKCDNPLDIGQYCPISLVGCLYKIV*K 370
>BI470619
Length = 422
Score = 32.7 bits (73), Expect = 0.50
Identities = 25/77 (32%), Positives = 35/77 (44%), Gaps = 8/77 (10%)
Frame = -2
Query: 465 TYEAASGQAINYRKSSIAFSRNTDSIGHHNITHLLGVVES--------MWHGKYLGLPSM 516
T+ A++G+ + K SI S NT IG HL G S + G ++ +PS+
Sbjct: 385 TFTASTGRGL---KPSIQLSNNTTVIGPSTSGHLEGSARSPLPLNADLSYLGNFIFIPSL 215
Query: 517 VERDKKSIFSFIKERIW 533
V K S S K R W
Sbjct: 214 VNLVKASRVSTDKARSW 164
>TC205454 similar to UP|LON1_ARATH (O64948) Lon protease homolog 1,
mitochondrial precursor , partial (25%)
Length = 1092
Score = 30.4 bits (67), Expect = 2.5
Identities = 17/45 (37%), Positives = 23/45 (50%)
Frame = +3
Query: 192 IWCWYFFFVITLDAFSKLTP*LFYMLQQQPNVKLPLYNKPFKLLL 236
I+C YFFF SK + * F + PN LP++ F +LL
Sbjct: 891 IFCKYFFFAFEQLYLSKFSK*CFSLSPSAPNPWLPVFC*SFVMLL 1025
>TC233646
Length = 978
Score = 30.0 bits (66), Expect = 3.2
Identities = 21/64 (32%), Positives = 36/64 (55%), Gaps = 1/64 (1%)
Frame = -3
Query: 256 PNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVL-ANRLKQILHKCISDSQAAF 314
P++I ++ LI KID + +K +SLCNV ++LS+ K I+ +Q +F
Sbjct: 841 PSII--SLVLIPKIDNLEILK*Y*SISLCNVTTRLLSRSS*PTSSKSFPS*PIARNQCSF 668
Query: 315 VPSR 318
+PS+
Sbjct: 667 IPSK 656
>TC205793 homologue to UP|Q9FLK9 (Q9FLK9) Leucine-rich repeat disease
resistance protein-like (Cf-5 disease resistance
protein-like), partial (21%)
Length = 334
Score = 29.3 bits (64), Expect = 5.5
Identities = 15/51 (29%), Positives = 25/51 (48%)
Frame = +3
Query: 297 NRLKQILHKCISDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIAL 347
N + I + ++A F F+L +T +LH CKT ++GN+ L
Sbjct: 12 NNERTISRSALKMARAPFTSFPFVL---ITLLSILHLSHCKTLKRDGNLKL 155
>BI315739
Length = 442
Score = 28.5 bits (62), Expect = 9.4
Identities = 15/42 (35%), Positives = 20/42 (46%)
Frame = +3
Query: 398 PFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQL 439
P SPYL+ IC E L I+ N +I R P I+ +
Sbjct: 48 PLSPYLF*ICMEKLAILIQSKVNEETWEPIKISRNGPGISPI 173
>TC214372
Length = 1309
Score = 28.5 bits (62), Expect = 9.4
Identities = 15/44 (34%), Positives = 22/44 (49%), Gaps = 1/44 (2%)
Frame = +1
Query: 38 LHGSTCKVMEFNVHQLVLPVIMIWK-MRCICLCTGSLLLIAGER 80
LH S CK F H + +P + IW M+ L G ++ G+R
Sbjct: 754 LHLSICKFHLFYHHCIYMPCVFIWMVMQSAKLFVGDAIMFNGDR 885
>TC206555 similar to UP|U315_ARATH (Q8LFJ5) UPF0315 protein At1g22270,
partial (90%)
Length = 621
Score = 28.5 bits (62), Expect = 9.4
Identities = 12/34 (35%), Positives = 18/34 (52%)
Frame = -2
Query: 521 KKSIFSFIKERIWKNIQNWSAWSLSRAGKEVLLK 554
+ S F +K+ IW + +W A S RA K+ K
Sbjct: 596 QNSDFVLVKKHIWNALTDWKATSGLRANKDTFFK 495
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.342 0.149 0.502
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,821,094
Number of Sequences: 63676
Number of extensions: 574768
Number of successful extensions: 4552
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 4482
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4546
length of query: 625
length of database: 12,639,632
effective HSP length: 103
effective length of query: 522
effective length of database: 6,081,004
effective search space: 3174284088
effective search space used: 3174284088
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC142224.5


