

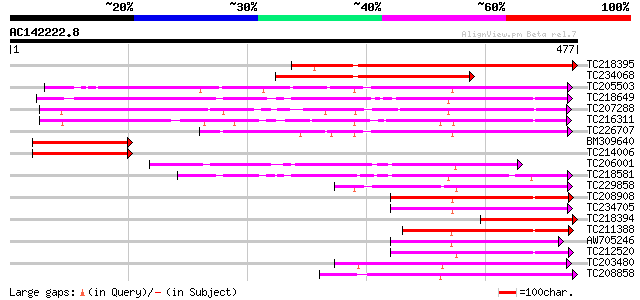
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.8 - phase: 0
(477 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like pr... 249 2e-66
TC234068 similar to GB|AAQ56818.1|34098871|BT010375 At3g03090 {A... 213 1e-55
TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete 207 6e-54
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 177 1e-44
TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete 147 8e-36
TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete 138 5e-33
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 138 5e-33
BM309640 similar to GP|9759246|dbj| sugar transporter-like prote... 126 2e-29
TC214006 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like pr... 125 3e-29
TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, parti... 118 5e-27
TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F2... 116 3e-26
TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporte... 109 3e-24
TC208908 weakly similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-... 108 5e-24
TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solu... 106 3e-23
TC218394 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like pr... 104 8e-23
TC211388 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like pr... 104 8e-23
AW705246 98 7e-21
TC212520 93 2e-19
TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 93 3e-19
TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%) 92 5e-19
>TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like protein,
partial (40%)
Length = 1013
Score = 249 bits (635), Expect = 2e-66
Identities = 132/242 (54%), Positives = 173/242 (70%), Gaps = 2/242 (0%)
Frame = +1
Query: 238 TFHDSAPQQVDEIMAEFS--YLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDH 295
T + +Q++E + Y +E++ E+F+G KA +I GLVLFQQI T
Sbjct: 1 TSDKESEKQIEETLVSLKSVYADQESEGNFLEVFQGPNLKAFIIGGGLVLFQQI---TGQ 171
Query: 296 GPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGI 355
+ GFS A+DAT+VS+++G+FKL+MT +AV+ VD LGRRPLL+GGVSGI
Sbjct: 172 PSVLYYAGPILQSAGFSAASDATKVSVVIGLFKLLMTWIAVLKVDDLGRRPLLIGGVSGI 351
Query: 356 VISLFLLGSYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIA 415
+SL LL +YY FL ++AV LLLYVGCYQISFGP+ WLM++E+FPLR RGKG+S+A
Sbjct: 352 ALSLVLLSAYYKFLGGFPLVAVGALLLYVGCYQISFGPISWLMVSEVFPLRTRGKGISLA 531
Query: 416 VLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAK 475
VL NFA+NA+VTFAFSPLK+ LGA LF +F AIA SL+FI F VPETKG++LE+IE+K
Sbjct: 532 VLTNFASNAVVTFAFSPLKEFLGAENLFLLFGAIATLSLLFIIFSVPETKGMSLEDIESK 711
Query: 476 CL 477
L
Sbjct: 712 IL 717
Score = 29.3 bits (64), Expect = 4.0
Identities = 28/112 (25%), Positives = 47/112 (41%), Gaps = 8/112 (7%)
Frame = +1
Query: 67 LTSGSLYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAP--------NFPLL 118
L G + G + VL FLG + V AL+ VG +F P FPL
Sbjct: 325 LLIGGVSGIALSLVLLSAYYKFLGGFPLVAVGALLLYVGCYQISFGPISWLMVSEVFPLR 504
Query: 119 VIGRLVFGIGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGL 170
G+ GI + + + A + A +P++ + + + F++ G +A L
Sbjct: 505 TRGK---GISLAVLTNFASNAVVTFAFSPLK-EFLGAENLFLLFGAIATLSL 648
>TC234068 similar to GB|AAQ56818.1|34098871|BT010375 At3g03090 {Arabidopsis
thaliana;} , partial (32%)
Length = 496
Score = 213 bits (543), Expect = 1e-55
Identities = 117/168 (69%), Positives = 130/168 (76%)
Frame = +1
Query: 224 KDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGEMFRGKCRKALVISAGL 283
KD AIRSLCQL+G+ F+DS P QVD I+AE SYLGEE + T GE+F+GKC KAL I +GL
Sbjct: 1 KDIAIRSLCQLRGQAFYDSVPWQVD*ILAELSYLGEEKEATFGELFQGKCLKALWIGSGL 180
Query: 284 VLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLG 343
VLFQQI T GFS A+DATRVSILLG FKLIMTGVAVVVVD+LG
Sbjct: 181 VLFQQI---TGQPSVLYYAGSIFQSAGFSGASDATRVSILLGFFKLIMTGVAVVVVDKLG 351
Query: 344 RRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAVVGLLLYVGCYQISF 391
RRPLLLGGVSGIVISLF LGSY I LDN+ V+AV+GLLLYVG YQISF
Sbjct: 352 RRPLLLGGVSGIVISLFFLGSYNIILDNSPVVAVIGLLLYVGSYQISF 495
>TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete
Length = 1928
Score = 207 bits (528), Expect = 6e-54
Identities = 145/466 (31%), Positives = 231/466 (49%), Gaps = 22/466 (4%)
Frame = +1
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
+L GYDIG S A I I+ + +EI LL +LY +LIGS LA +D++
Sbjct: 292 ILLGYDIGVMSGAAIYIKRDLKVSD-----EQIEI-LLGIINLY-SLIGSCLAGRTSDWI 450
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR + + ++LVG+ + F P++ L+ GR V GIGIG A+ AP+Y AE +P R
Sbjct: 451 GRRYTIGLGGAIFLVGSTLMGFYPHYSFLMCGRFVAGIGIGYALMIAPVYTAEVSPASSR 630
Query: 150 GQLVSLKEFF----IVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
G L S E F I++G ++ YG L + GWR M G+ + +V++ G+ +P S
Sbjct: 631 GFLTSFPEVFINGGILLGYISNYGFSKLTLK--VGWRMMLGVGAIPSVVLTEGVLAMPES 804
Query: 206 PRWILLR--------AIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYL 257
PRW+++R + K D + + + Q G +S V ++ + +
Sbjct: 805 PRWLVMRGRLGEARKVLNKTSDSKEEAQLRLAEIKQAAG--IPESCNDDVVQVNKQSNGE 978
Query: 258 GEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLA 314
G ++ L R ++ + G+ FQQ + V + P R G +
Sbjct: 979 GVWKELFLYP--TPAIRHIVIAALGIHFFQQASGVDAVVLYSP------RIFEKAGITND 1134
Query: 315 ADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN--- 371
++ +G K + A +DR+GRRPLLL V G+V+SL L +D+
Sbjct: 1135THKLLATVAVGFVKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLAISLTVIDHSER 1314
Query: 372 ----AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVT 427
A ++ +L YV + I GP+ W+ +EIFPLRLR +G + V VN +A+V+
Sbjct: 1315KLMWAVGSSIAMVLAYVATFSIGAGPITWVYSSEIFPLRLRAQGAAAGVAVNRTTSAVVS 1494
Query: 428 FAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
F L + G F+++ IA +F Y ++PET+G TLE++E
Sbjct: 1495MTFLSLTRAITIGGAFFLYCGIATVGWIFFYTVLPETRGKTLEDME 1632
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 177 bits (448), Expect = 1e-44
Identities = 128/461 (27%), Positives = 232/461 (49%), Gaps = 10/461 (2%)
Frame = +2
Query: 23 LFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLA 82
L A G + FG+ G TS +I I L E L S S GA++G++ +
Sbjct: 188 LIVALGPIQFGFTAGYTSPTQSAI-------INDLGLSVSEFSLFGSLSNVGAMVGAIAS 346
Query: 83 FNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAE 142
IA+++GR+ L++A++ ++G L +FA + L +GRL+ G G+G+ + P+YIAE
Sbjct: 347 GQIAEYIGRKGSLMIASIPNIIGWLAISFAKDSSFLYMGRLLEGFGVGIISYTVPVYIAE 526
Query: 143 TAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWL 202
+P +RG LVS+ + + IGI+ Y LG + WR + I I+ ++++
Sbjct: 527 ISPPNLRGGLVSVNQLSVTIGIMLAYLLGIFV-----EWRILAIIGILPCTILIPALFFI 691
Query: 203 PASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEND 262
P SPRW+ + ++ +T+++ L R F +V+EI +
Sbjct: 692 PESPRWLAKMGMTEE------FETSLQVL-----RGFDTDISVEVNEIKRAVASTNTRIT 838
Query: 263 VTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSI 322
V ++ + + L+I GL++ QQ+ + ++ R G S ++DA +
Sbjct: 839 VRFADLKQRRYWLPLMIGIGLLILQQLSGINGVLFYSSTIFR---NAGIS-SSDAATFGV 1006
Query: 323 LLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLG-SYYI---------FLDNA 372
G +++ T + + + D+ GRR LL+ +G+ SL ++ ++YI
Sbjct: 1007--GAVQVLATSLTLWLADKSGRRLLLIVSATGMSFSLLVVAITFYIKASISETSSLYGIL 1180
Query: 373 AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
+ L++VG++ V + + G M W++++EI P+ ++G S+A L N+ + LVT +
Sbjct: 1181STLSLVGVVAMVIAFSLGMGAMPWIIMSEILPINIKGLAGSVATLANWLFSWLVTLTANM 1360
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
L D G F I++ + ++VF+ VPETKG T+EEI+
Sbjct: 1361LLDWSSGG-TFTIYAVVCALTVVFVTIWVPETKGKTIEEIQ 1480
>TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete
Length = 1813
Score = 147 bits (372), Expect = 8e-36
Identities = 131/480 (27%), Positives = 213/480 (44%), Gaps = 33/480 (6%)
Frame = +1
Query: 26 AFGGLLFGYDIGATSSA-------------TISIQSSSLSGITWYDLDAVEIGLLTSGSL 72
A GG LFGYD+G + + L + D + L TS
Sbjct: 127 ALGGSLFGYDLGVSGGVPSMDDFLKEFFPKVYRRKQMHLHETDYCKYDDQVLTLFTSSLY 306
Query: 73 YGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLA 132
+ AL+ + A + GR+ ++V AL +L GA++ A A N +L+IGR++ G GIG
Sbjct: 307 FSALVMTFFASFLTRKKGRKASIIVGALSFLAGAILNAAAKNIAMLIIGRVLLGGGIGFG 486
Query: 133 MHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTV--AGWRYMFGISSP 190
A P+Y++E AP RG + L +F GI+ L + + + GWR G++
Sbjct: 487 NQAVPLYLSEMAPAKNRGAVNQLFQFTTCAGILIA-NLVNYFTEKIHPYGWRISLGLAGL 663
Query: 191 VAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEI 250
A M G +P ++ ++G L D A + L +++G + +
Sbjct: 664 PAFAMLVGGICCAETP-----NSLVEQGRL----DKAKQVLQRIRG----------TENV 786
Query: 251 MAEFSYLGEENDVT------LGEMFRGKCRKALVISA-GLVLFQQI---QTVTDHGPTKC 300
AEF L E ++ + + K R L+I A G+ FQQ+ ++ + P
Sbjct: 787 EAEFEDLKEASEEAQAVKSPFRTLLKRKYRPQLIIGALGIPAFQQLTGNNSILFYAPV-- 960
Query: 301 SLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLF 360
GF A I G L+ T +++ +VD+ GRR L ++ +
Sbjct: 961 ----IFQSLGFGANASLFSSFITNGAL-LVATVISMFLVDKYGRRKFFLEAGFEMICCMI 1125
Query: 361 LLGSYYI--------FLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGL 412
+ G+ + VV + L+V Y S+GP+GWL+ +E+FPL +R
Sbjct: 1126ITGAVLAVNFGHGKEIGKGVSAFLVVVIFLFVLAYGRSWGPLGWLVPSELFPLEIRSSAQ 1305
Query: 413 SIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
SI V VN ALV F L GI F +F+++ + F++F++PETK + +EEI
Sbjct: 1306SIVVCVNMIFTALVAQLFLMSLCHLKFGI-FLLFASLIIFMSFFVFFLLPETKKVPIEEI 1482
>TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete
Length = 1925
Score = 138 bits (348), Expect = 5e-33
Identities = 130/486 (26%), Positives = 223/486 (45%), Gaps = 39/486 (8%)
Frame = +1
Query: 26 AFGGLLFGYDIGATSSAT------------ISIQSSSLSGITWY-DLDAVEIGLLTSGSL 72
A GGL+FGYDIG + T + + +S + Y D+ + + TS
Sbjct: 187 AMGGLIFGYDIGISGGVTSMDPFLLKFFPSVFRKKNSDKTVNQYCQYDSQTLTMFTSSLY 366
Query: 73 YGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLA 132
AL+ S++A + GR+ +L L++LVGALI FA + +L++GR++ G GIG A
Sbjct: 367 LAALLSSLVASTVTRRFGRKLSMLFGGLLFLVGALINGFAQHVWMLIVGRILLGFGIGFA 546
Query: 133 MHAAPMYIAETAPTPIRGQLVSLKEFFIVI--------GIVAGYGLGSLL-VDTVAGWRY 183
A PI + + + GI G + LL + ++ W +
Sbjct: 547 NQVC-------ATLPI*NGFIQI*RSIGTLAFNCQSQFGIPRGQCVELLLWLKSMVAWGW 705
Query: 184 MFGI---SSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFH 240
+ + A+I+ G LP +P ++ ++GD + K QLQ
Sbjct: 706 KIEVWEGAMVPALIITVGSLVLPDTPNSMI-----ERGDREKAK-------AQLQRIRGI 849
Query: 241 DSAPQQVDEIMAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGP 297
D+ ++ ++++A S + + + + K R L ++ + FQQ I + + P
Sbjct: 850 DNVDEEFNDLVAA-SESSSQVEHPWRNLLQRKYRPHLTMAVLIPFFQQLTGINVIMFYAP 1026
Query: 298 TKCSLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLL-GGVSGIV 356
S + GF A A +++ GV ++ T V++ VD+ GRR L L GGV ++
Sbjct: 1027VLFSSI------GFKDDA-ALMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGVQMLI 1185
Query: 357 ISLFL---LGSYYIFLDNA-------AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLR 406
+ +G+ + N A++ V+ + +YV + S+GP+GWL+ +EIFPL
Sbjct: 1186CQAVVAAAIGAKFGTDGNPGDLPKWYAIVVVLFICIYVSAFAWSWGPLGWLVPSEIFPLE 1365
Query: 407 LRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKG 466
+R SI V VN L+ F + + G LF F+ + F+YF +PETKG
Sbjct: 1366IRSAAQSINVSVNMLFTFLIAQVFLTMLCHMKFG-LFLFFAFFVLIMTFFVYFFLPETKG 1542
Query: 467 LTLEEI 472
+ +EE+
Sbjct: 1543IPIEEM 1560
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 138 bits (348), Expect = 5e-33
Identities = 97/329 (29%), Positives = 158/329 (47%), Gaps = 15/329 (4%)
Frame = +3
Query: 160 IVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGD 219
I+IG ++ YG L + GWR M G+ + ++++G + +P SPRW++ + +
Sbjct: 3 ILIGYISNYGFSKLALRL--GWRLMLGVGAIPSILIGVAVLAMPESPRWLVAKGRLGEAK 176
Query: 220 LQTLKDTAIRSLCQLQGRTFHDSA--PQQVDEIMAEFSYLGEENDVTLGEMF---RGKCR 274
K + +L+ D+A PQ D+ + S + V E+F R
Sbjct: 177 RVLYKISESEEEARLRLADIKDTAGIPQDCDDDVVLVSKQTHGHGVWR-ELFLHPTPAVR 353
Query: 275 KALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIM 331
+ S G+ F Q I V + P R G ++ +G K +
Sbjct: 354 HIFIASLGIHFFAQATGIDAVVLYSP------RIFEKAGIKSDNYRLLATVAVGFVKTVS 515
Query: 332 TGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN-------AAVLAVVGLLLYV 384
VA +DR GRR LLL VSG+++SL LG +D+ A L++ +L YV
Sbjct: 516 ILVATFFLDRAGRRVLLLCSVSGLILSLLTLGLSLTVVDHSQTTLNWAVGLSIAAVLSYV 695
Query: 385 GCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFY 444
+ I GP+ W+ +EIFPLRLR +G++I VN + ++ F L+ + G F+
Sbjct: 696 ATFSIGSGPITWVYSSEIFPLRLRAQGVAIGAAVNRVTSGVIAMTFLSLQKAITIGGAFF 875
Query: 445 IFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+F+ +A + +F Y ++PET+G TLEEIE
Sbjct: 876 LFAGVAAVAWIFHYTLLPETRGKTLEEIE 962
>BM309640 similar to GP|9759246|dbj| sugar transporter-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 390
Score = 126 bits (317), Expect = 2e-29
Identities = 59/84 (70%), Positives = 75/84 (89%)
Frame = +3
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L FLFPA GGLLFGYDIGATS ATIS+QS LSGI+W++L A+++GL+ SGSLYGAL+GS
Sbjct: 135 LPFLFPALGGLLFGYDIGATSGATISLQSPELSGISWFNLSAIQLGLVVSGSLYGALLGS 314
Query: 80 VLAFNIADFLGRRRELLVAALMYL 103
++AF IADFLGR+++L+ AAL+YL
Sbjct: 315 LVAFAIADFLGRKKQLITAALLYL 386
>TC214006 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like protein,
partial (17%)
Length = 595
Score = 125 bits (315), Expect = 3e-29
Identities = 59/84 (70%), Positives = 74/84 (87%)
Frame = +3
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L FLFPA GGLLFGYDIGATS ATIS+QS LSGI+W+ L A+++GL+ SGSLYGAL+GS
Sbjct: 339 LPFLFPALGGLLFGYDIGATSGATISLQSPELSGISWFKLSAIQLGLVVSGSLYGALLGS 518
Query: 80 VLAFNIADFLGRRRELLVAALMYL 103
++AF IADFLGR+++L+ AAL+YL
Sbjct: 519 LVAFAIADFLGRKKQLITAALLYL 590
>TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, partial (66%)
Length = 951
Score = 118 bits (296), Expect = 5e-27
Identities = 85/320 (26%), Positives = 149/320 (46%), Gaps = 6/320 (1%)
Frame = +3
Query: 118 LVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDT 177
L +GR G GIG+ + P+YIAE AP +RG L + + IV G G S L+ +
Sbjct: 39 LDMGRFFTGYGIGVISYVVPVYIAEIAPKNLRGGLATTNQLLIVTG-----GSVSFLLGS 203
Query: 178 VAGWRYMFGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGR 237
V WR + + + G+ ++P SPRW+ +K+ L +L +L+G+
Sbjct: 204 VINWRELALAGLVPCICLLVGLCFIPESPRWLAKVGREKEFQL---------ALSRLRGK 356
Query: 238 TFHDSAPQQVDEIMAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGP 297
H + EI+ L L ++F+ K ++VI GL+ QQ + G
Sbjct: 357 --HADISDEAAEILDYIETLESLPKTKLLDLFQSKYVHSVVIGVGLMACQQSVGINGIGF 530
Query: 298 TKCSLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVI 357
+ G S T I ++ T + +++D+ GRRPL++ +G +
Sbjct: 531 YTAEIFVA---AGLSSGKAGT---IAYACIQIPFTLLGAILMDKSGRRPLVMVSAAGTFL 692
Query: 358 SLFLLGSYYIFLDNAA------VLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKG 411
F+ + D + +LA G+L+Y+ + I G + W++++EIFP+ L+G
Sbjct: 693 GCFVAAFAFFLKDQSLLPEWVPILAFAGVLIYIAAFSIGLGSVPWVIMSEIFPIHLKGTA 872
Query: 412 LSIAVLVNFAANALVTFAFS 431
S+ VLV + +V++ F+
Sbjct: 873 GSLVVLVAWLGAWVVSYTFN 932
>TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;} , partial (66%)
Length = 1340
Score = 116 bits (290), Expect = 3e-26
Identities = 96/345 (27%), Positives = 173/345 (49%), Gaps = 13/345 (3%)
Frame = +2
Query: 142 ETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWW 201
E AP + G L S+ + I IGI+ Y LG + WR + + ++ G+++
Sbjct: 5 EIAPNHLIGGLGSVNQLSITIGIMLAYLLGLFV-----NWRILAILGILPCTVLIPGLFF 169
Query: 202 LPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEN 261
+P SPRW+ K G + +T+++ L R F +V EI + G+
Sbjct: 170 IPESPRWLA-----KMGMIDEF-ETSLQVL-----RGFDTDISVEVHEIKRSVASTGKRA 316
Query: 262 DVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVS 321
+ ++ R + L++ GL++ QQ+ + +G S N G S + AT
Sbjct: 317 AIRFADLKRKRYWFPLMVGIGLLVLQQLSGI--NGILFYSTTIFAN-AGISSSEAAT--- 478
Query: 322 ILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLG-SYYI---------FLDN 371
+ LG ++I TG++ +VD+ GRR LL+ S + +SL ++ ++Y+
Sbjct: 479 VGLGAVQVIATGISTWLVDKSGRRLLLIISSSVMTVSLLIVSIAFYLEGVVSEDSHLFSI 658
Query: 372 AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFS 431
++++VGL+ V + + GP+ WL+++EI P+ ++G SIA + N+ L+++ +
Sbjct: 659 LGIVSIVGLVAMVIGFSLGLGPIPWLIMSEILPVNIKGLAGSIATMGNW----LISWGIT 826
Query: 432 PLKDLL---GAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+LL +G F I++ +A ++ FI VPETKG TLEEI+
Sbjct: 827 MTANLLLNWSSGGTFTIYTVVAAFTIAFIAMWVPETKGRTLEEIQ 961
>TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporter 10 (Mus
musculus 13 days embryo heart cDNA, RIKEN full-length
enriched library, clone:D330001H08 product:solute
carrier family 2 (facilitated glucose transporter),
member 10, full insert sequence), partial (4%)
Length = 715
Score = 109 bits (272), Expect = 3e-24
Identities = 68/210 (32%), Positives = 109/210 (51%), Gaps = 10/210 (4%)
Frame = +3
Query: 274 RKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLI 330
R A ++ AGL+ FQQ I TV + PT + GF A +S+++
Sbjct: 9 RLAFLVGAGLLAFQQFTGINTVMYYSPT------IVQMAGFHANELALLLSLIVAGMNAA 170
Query: 331 MTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAV-------LAVVGLLLY 383
T + + ++D GR+ L L + G+++SL +L + F ++ LAVVGL LY
Sbjct: 171 GTILGIYLIDHAGRKKLALSSLGGVIVSLVILA--FAFYKQSSTSNELYGWLAVVGLALY 344
Query: 384 VGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILF 443
+G + GP+ W + +EI+P RG ++ V + +N +V+ F + + +G G F
Sbjct: 345 IGFFSPGMGPVPWTLSSEIYPEEYRGICGGMSATVCWVSNLIVSETFLSIAEGIGIGSTF 524
Query: 444 YIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
I IAV + VF+ VPETKGLT +E+E
Sbjct: 525 LIIGVIAVVAFVFVLVYVPETKGLTFDEVE 614
>TC208908 weakly similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like
protein, partial (28%)
Length = 753
Score = 108 bits (270), Expect = 5e-24
Identities = 58/160 (36%), Positives = 100/160 (62%), Gaps = 6/160 (3%)
Frame = +2
Query: 321 SILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN------AAV 374
+I + K+ MT + V+++D+ GRRPLLL G + FL ++ D + +
Sbjct: 2 TIAIVAVKIPMTTIGVLLMDKSGRRPLLLVSAVGTCVGCFLAALSFVLQDLHKWKGVSPI 181
Query: 375 LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLK 434
LA+VG+L+YVG Y I G + W++++EIFP+ ++G S+ LV++ + +++++F+ L
Sbjct: 182 LALVGVLVYVGSYSIGMGAIPWVIMSEIFPINVKGSAGSLVTLVSWLCSWIISYSFNFLM 361
Query: 435 DLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
AG F +FS+I +++F+ +VPETKG TLEEI+A
Sbjct: 362 SWSSAG-TFLMFSSICGFTVLFVAKLVPETKGRTLEEIQA 478
>TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solute carrier
family 2 (facilitated glucose transporter), member 12
{Mus musculus;} , partial (5%)
Length = 992
Score = 106 bits (264), Expect = 3e-23
Identities = 51/156 (32%), Positives = 94/156 (59%), Gaps = 3/156 (1%)
Frame = +1
Query: 321 SILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN---AAVLAV 377
++ +GV K I VA++++D+LGR+PLL+ G+ + LF +G+ L A LA+
Sbjct: 55 TVAVGVAKTIFILVAIILIDKLGRKPLLMISTIGMTVCLFCMGATLALLGKGSFAIALAI 234
Query: 378 VGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLL 437
+ + V + + GP+ W++ +EIFPLR+R + ++ + N + LV +F + + +
Sbjct: 235 LFVCGNVAFFSVGLGPVCWVLTSEIFPLRVRAQASALGAVANRVCSGLVAMSFLSVSEAI 414
Query: 438 GAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
F++F+AI+ ++ F+ +VPETKG +LE+IE
Sbjct: 415 SVAGTFFVFAAISALAIAFVVTLVPETKGKSLEQIE 522
>TC218394 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like protein,
partial (15%)
Length = 762
Score = 104 bits (260), Expect = 8e-23
Identities = 51/81 (62%), Positives = 65/81 (79%)
Frame = +3
Query: 397 LMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVF 456
LM++E P R+RGKG+ +AV NFA+NA+VTFAFSPLK+ LGA LF +F AIA SL+F
Sbjct: 12 LMVSEXXPXRIRGKGIXLAVXTNFASNAVVTFAFSPLKEFLGAENLFLLFGAIATLSLLF 191
Query: 457 IYFIVPETKGLTLEEIEAKCL 477
I F VPETKG++LE+IE+K L
Sbjct: 192 IIFSVPETKGMSLEDIESKIL 254
>TC211388 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like protein,
partial (27%)
Length = 786
Score = 104 bits (260), Expect = 8e-23
Identities = 55/150 (36%), Positives = 93/150 (61%), Gaps = 6/150 (4%)
Frame = +3
Query: 331 MTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLD------NAAVLAVVGLLLYV 384
MT + V+++D+ GRRPLLL SG + FL + D + +LA+ G+L+Y
Sbjct: 93 MTALGVLLMDKSGRRPLLLISASGTCLGCFLAALSFTLQDLHKWKEGSPILALAGVLVYT 272
Query: 385 GCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFY 444
G + + G + W++++EIFP ++G S+ LV++ + +V++AF+ L AG F+
Sbjct: 273 GSFSLGMGGIPWVIMSEIFPTNVKGSAGSLVTLVSWLCSWIVSYAFNFLMSWSSAG-TFF 449
Query: 445 IFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
IFS+I +++F+ +VPETKG TLEE++A
Sbjct: 450 IFSSICGFTILFVAKLVPETKGRTLEEVQA 539
>AW705246
Length = 484
Score = 98.2 bits (243), Expect = 7e-21
Identities = 55/153 (35%), Positives = 86/153 (55%), Gaps = 7/153 (4%)
Frame = +2
Query: 321 SILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLD------NAAV 374
S+ +G K + VA + +DR GRR LLL VSG+++SL LG +D N AV
Sbjct: 11 SVAVGFDKTVCIWVATIFLDRAGRRVLLLCNVSGLILSLMTLGLSLTVVDHSQTTLNCAV 190
Query: 375 -LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPL 433
L + +L YV + I GP+ W+ I+EIFPLRL G++I VN + ++ F L
Sbjct: 191 GLRIDSVLSYVATFSIWSGPITWVYISEIFPLRLWAHGVAIGAAVNKVTSGVIAMTFLYL 370
Query: 434 KDLLGAGILFYIFSAIAVASLVFIYFIVPETKG 466
+ + G F++F+ +A + +F Y ++PE +G
Sbjct: 371 QKAITIGGAFFLFAGVAAVAWIFHYTLIPEARG 469
>TC212520
Length = 720
Score = 93.2 bits (230), Expect = 2e-19
Identities = 50/160 (31%), Positives = 92/160 (57%), Gaps = 6/160 (3%)
Frame = +3
Query: 321 SILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISL-FLLGSYYIFLDNAAV----- 374
+I +++ G+ ++D+ GR+PL+L SG+V F+ ++Y+ + V
Sbjct: 63 TITYACLQIVXXGLGAALIDKAGRKPLILLSGSGLVAGCTFVAVAFYLKVHEVGVEAVPA 242
Query: 375 LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLK 434
LAV G+L+Y+G + I G + W+++ EIFP+ ++G S+A LVN+ L ++ F+
Sbjct: 243 LAVTGILVYIGSFSIGMGAIPWVVMCEIFPVNIKGLAGSVATLVNWFGAWLCSYTFNFFM 422
Query: 435 DLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
G F +++AI ++FI VPE+KG +LE+++A
Sbjct: 423 SWSSYG-TFILYAAINALDILFIIVAVPESKGKSLEQLQA 539
>TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (38%)
Length = 1040
Score = 92.8 bits (229), Expect = 3e-19
Identities = 60/208 (28%), Positives = 111/208 (52%), Gaps = 9/208 (4%)
Frame = +2
Query: 274 RKALVISAGLVLFQQIQTV------TDHGPTKCSLLRCINPTGFSLAADATRVSILLGVF 327
+ ALV+ G+ + QQ + T + + ++ G + + +S
Sbjct: 200 KHALVVGVGIQILQQFSGINGVLYYTPQILEEAGVEVLLSDIGIGSESASFLISAFTTFL 379
Query: 328 KLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLL--GSYYIFLDNA-AVLAVVGLLLYV 384
L GVA+ ++D GRR LLL + +++SL +L GS F + A A ++ V +++Y
Sbjct: 380 MLPCIGVAMKLMDVSGRRQLLLTTIPVLIVSLIILVIGSLVNFGNVAHAAISTVCVVVYF 559
Query: 385 GCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFY 444
C+ + +GP+ ++ +EIFP R+RG ++I LV + + ++T++ + LG G +F
Sbjct: 560 CCFVMGYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDIIITYSLPVMLGSLGLGGVFA 739
Query: 445 IFSAIAVASLVFIYFIVPETKGLTLEEI 472
I++ + S +F++ VPETKG+ LE I
Sbjct: 740 IYAVVCFISWIFVFLKVPETKGMPLEVI 823
>TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%)
Length = 993
Score = 92.0 bits (227), Expect = 5e-19
Identities = 65/221 (29%), Positives = 117/221 (52%), Gaps = 4/221 (1%)
Frame = +3
Query: 261 NDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGF-SLAADATR 319
+ V L E+ G+ + + I + L QQ+ + + + + T F S +
Sbjct: 102 DSVKLSELIYGRYFRVMFIGSTLFALQQLSGI--------NAVFYFSSTVFESFGVPSDI 257
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFL---LGSYYIFLDNAAVLA 376
+ +GV L+ + VA++++D+LGR+ LLLG G+ +S+ L S + + L+
Sbjct: 258 ANSCVGVCNLLGSVVAMILMDKLGRKVLLLGSFLGMGLSMGLQVIAASSFASGFGSMYLS 437
Query: 377 VVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDL 436
V G+LL+V + GP+ L+++EI P +R K ++I + V++ N V F L +L
Sbjct: 438 VGGMLLFVLSFAFGAGPVPCLIMSEILPSNIRAKAMAICLAVHWVINFFVGLFFLRLLEL 617
Query: 437 LGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
+GA +L+ IF + ++VF+ + ETKG +L+EIE L
Sbjct: 618 IGAQLLYSIFGFCCLIAVVFVKKNILETKGKSLQEIEIALL 740
Score = 28.1 bits (61), Expect = 9.0
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 9/89 (10%)
Frame = +3
Query: 76 LIGSVLAFNIADFLGRRRELLVAAL-----MYLVGALITAFAPNFP---LLVIGRLVFGI 127
L+GSV+A + D LGR+ LL + L M L ++FA F L V G L+F +
Sbjct: 285 LLGSVVAMILMDKLGRKVLLLGSFLGMGLSMGLQVIAASSFASGFGSMYLSVGGMLLFVL 464
Query: 128 GIGLAMHAAP-MYIAETAPTPIRGQLVSL 155
P + ++E P+ IR + +++
Sbjct: 465 SFAFGAGPVPCLIMSEILPSNIRAKAMAI 551
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.328 0.145 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,670,453
Number of Sequences: 63676
Number of extensions: 295482
Number of successful extensions: 2023
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 1952
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1969
length of query: 477
length of database: 12,639,632
effective HSP length: 101
effective length of query: 376
effective length of database: 6,208,356
effective search space: 2334341856
effective search space used: 2334341856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC142222.8


