

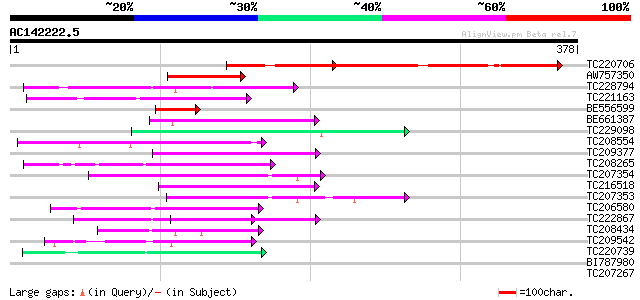
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.5 + phase: 0
(378 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220706 311 2e-85
AW757350 74 1e-13
TC228794 similar to UP|Q6V8V0 (Q6V8V0) Transducin-like (Fragment... 67 1e-11
TC221163 61 9e-10
BE556599 similar to GP|22831203|dbj P0496C02.23 {Oryza sativa (j... 60 2e-09
BE661387 54 2e-07
TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory pr... 52 6e-07
TC208554 UP|Q9FUF3 (Q9FUF3) FAS2, complete 51 1e-06
TC209377 similar to UP|Q9FLX9 (Q9FLX9) Notchless protein homolog... 50 1e-06
TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%) 50 2e-06
TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 48 8e-06
TC216518 weakly similar to UP|Q9LW87 (Q9LW87) Coatomer protein c... 47 1e-05
TC207353 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 47 1e-05
TC206580 weakly similar to UP|Q9JKK6 (Q9JKK6) G beta-like protei... 46 3e-05
TC222867 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Frag... 45 5e-05
TC208434 similar to PIR|T01391|T01391 WD-repeat protein T4I9.10 ... 45 5e-05
TC209542 43 2e-04
TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nu... 42 3e-04
BI787980 similar to SP|P25387|GBLP Guanine nucleotide-binding pr... 40 0.001
TC207267 similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (53%) 40 0.001
>TC220706
Length = 891
Score = 311 bits (798), Expect = 2e-85
Identities = 165/224 (73%), Positives = 179/224 (79%)
Frame = +2
Query: 145 TFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIM 204
TF +K S S C +R EFSEDELLSVVLMKNGRKVVCGSQTG++
Sbjct: 71 TFLPFFIKTTDNSSSSENSSCRIRHE------EFSEDELLSVVLMKNGRKVVCGSQTGVI 232
Query: 205 LLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEH 264
LLYSWGCFKDCSDRF DLSSNSID MLKLDEDRIITGSENG+INLVGILPNRVIQP+AEH
Sbjct: 233 LLYSWGCFKDCSDRFTDLSSNSIDAMLKLDEDRIITGSENGIINLVGILPNRVIQPIAEH 412
Query: 265 SEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDNILQGSRNTQRNENGGVDNDVD 324
SEYPVE L AFSHD+KFLGSIAHDQMLKLWDLDNILQGS NTQRNE GG VD
Sbjct: 413 SEYPVECL------AFSHDKKFLGSIAHDQMLKLWDLDNILQGSGNTQRNEAGGA---VD 565
Query: 325 SDNEDEMDVDNNTSKFTKGNKRKNASKGHAAGDSNNFFADL*EI 368
SD +DEMD+D++ SK KGNKRKNA+ G+A G SNNFFADL*E+
Sbjct: 566 SD-DDEMDLDDDPSKINKGNKRKNANNGNALGGSNNFFADL*EV 694
Score = 89.7 bits (221), Expect = 2e-18
Identities = 46/74 (62%), Positives = 50/74 (67%)
Frame = -1
Query: 145 TFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIM 204
TF +K S S C +R EFSEDELLSVVLMKNGRKVVCGS TG++
Sbjct: 204 TFLPFFIKTTDNSSSSENSSCRIRHE------EFSEDELLSVVLMKNGRKVVCGSXTGVI 43
Query: 205 LLYSWGCFKDCSDR 218
LLYSWGCFKDCSDR
Sbjct: 42 LLYSWGCFKDCSDR 1
>AW757350
Length = 157
Score = 73.9 bits (180), Expect = 1e-13
Identities = 35/52 (67%), Positives = 42/52 (80%)
Frame = +2
Query: 106 LTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATS 157
LTEST A GDD+GC+KVWD ER+C NSF A+ED I ++TF SDAM L+ATS
Sbjct: 2 LTESTGA*GDDEGCLKVWDAIERTCSNSFNANEDDIYNMTFVSDAMNLVATS 157
>TC228794 similar to UP|Q6V8V0 (Q6V8V0) Transducin-like (Fragment), complete
Length = 870
Score = 67.4 bits (163), Expect = 1e-11
Identities = 46/185 (24%), Positives = 89/185 (47%), Gaps = 2/185 (1%)
Frame = +3
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + F+P+ ++VA+G D ++ L+ N + D + + H + + G +
Sbjct: 291 YTMKFNPAGSVVASGSHDREIFLW-----NVHGDCKNFMVLKGHKNAVLDLHWTTDGTQI 455
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES--TVASGDDDGCIKVWDTRE 127
++ SPD ++ A DVETG I ++ H + VN V SG DDG K+WD R+
Sbjct: 456 VSASPDKTVRAWDVETGKQIKKMVE-HLSYVNSCCPSRRGPPLVVSGSDDGTAKLWDMRQ 632
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVV 187
R +F + I+ + F+ + K+ D + + +LR+ +V + +D + ++
Sbjct: 633 RGSIQTF-PDKYQITAVGFSDASDKIFTGGIDNDVKIWDLRKGEVTMTLQGHQDMITAMQ 809
Query: 188 LMKNG 192
L +G
Sbjct: 810 LSPDG 824
>TC221163
Length = 832
Score = 60.8 bits (146), Expect = 9e-10
Identities = 39/150 (26%), Positives = 68/150 (45%)
Frame = +3
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ + + L +G DG +H+ ++ + + +H++S F G
Sbjct: 222 LTINSTSTLALSGSKDGSVHIVNITTGRVVDNNA----LASHSDSIECVGFAPSGSWAAV 389
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCC 131
G D ++ D+E + R HE V L L S VASG DG +++WD+R C
Sbjct: 390 GGMDKKLIIWDIE--HLLPRGTCEHEDGVTCLAWLGASYVASGCVDGKVRLWDSRSGECV 563
Query: 132 NSFEAHEDYISDITFASDAMKLLATSGDGT 161
+ + H D I ++ +S+ L++ S DGT
Sbjct: 564 KTLKGHSDAIQSLSVSSNRNYLVSASVDGT 653
Score = 36.6 bits (83), Expect = 0.019
Identities = 43/204 (21%), Positives = 83/204 (40%), Gaps = 2/204 (0%)
Frame = +3
Query: 116 DDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQ 175
+D I +W+T + N+F H D ++ F D + S D TL + N +
Sbjct: 3 EDFSIWMWNTDNAALLNTFIGHGDSVTCGDFTPDGKIICTGSDDATLRIWNPK------- 161
Query: 176 SEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDE 235
G T ++ + + ++ L+ NS T+
Sbjct: 162 ---------------------TGESTHVVRGHPYH-----TEGLTCLTINSTSTL----- 248
Query: 236 DRIITGSENGMINLVGILPNRVI--QPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHD 293
++GS++G +++V I RV+ +A HS+ +E +G F+ + D
Sbjct: 249 --ALSGSKDGSVHIVNITTGRVVDNNALASHSD-SIECVG------FAPSGSWAAVGGMD 401
Query: 294 QMLKLWDLDNILQGSRNTQRNENG 317
+ L +WD++++L R T +E+G
Sbjct: 402 KKLIIWDIEHLL--PRGTCEHEDG 467
>BE556599 similar to GP|22831203|dbj P0496C02.23 {Oryza sativa (japonica
cultivar-group)}, partial (7%)
Length = 167
Score = 60.1 bits (144), Expect = 2e-09
Identities = 26/30 (86%), Positives = 29/30 (96%)
Frame = +2
Query: 98 AAVNRLVNLTESTVASGDDDGCIKVWDTRE 127
AA+NRL+NLTESTVASGDD+GCIKVWD RE
Sbjct: 77 AAINRLINLTESTVASGDDEGCIKVWDIRE 166
>BE661387
Length = 636
Score = 53.5 bits (127), Expect = 2e-07
Identities = 28/116 (24%), Positives = 59/116 (50%), Gaps = 3/116 (2%)
Frame = +1
Query: 94 NAHEAAVNRLVNLT---ESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDA 150
+AH+ VN + T + + +G DD KVWD + +SC + E H +S + F +
Sbjct: 67 DAHQKGVNCVDYFTGGDKPYLITGSDDHTAKVWDYQTKSCVQTLEGHTHNVSAVCFHPEL 246
Query: 151 MKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLL 206
++ S DGT+ + + +++ + + + ++ +K+ R+VV G G +++
Sbjct: 247 PIIITGSEDGTVRIWHSTTYRLENTLNYGLERVWAIGYLKSSRRVVIGYDEGTIMV 414
>TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (59%)
Length = 580
Score = 51.6 bits (122), Expect = 6e-07
Identities = 43/188 (22%), Positives = 74/188 (38%), Gaps = 3/188 (1%)
Frame = +3
Query: 82 DVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYI 141
DV TGS I L N + + SG D ++VWD + C AH D +
Sbjct: 12 DVPTGSLIKTLHGHTNYVFCVNFNPQSNIIVSGSFDETVRVWDVKSGKCLKVLPAHSDPV 191
Query: 142 SDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVV-LMKNGRKVVCGSQ 200
+ + F D ++++S DG + + ++ +S V N + ++ G+
Sbjct: 192 TAVDFNRDGSLIVSSSYDGLCRIWDASTGHCMKTLIDDDNPPVSFVKFSPNAKFILVGTL 371
Query: 201 TGIMLL--YSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGILPNRVI 258
+ L YS G F V+ T + I+ GSE I L + +++
Sbjct: 372 DNTLRLWNYSTGKFLKTYTGHVNSKYCISSTFSTTNGKYIVGGSEENYIYLWDLQSRKIV 551
Query: 259 QPVAEHSE 266
Q + HS+
Sbjct: 552 QKLEGHSD 575
>TC208554 UP|Q9FUF3 (Q9FUF3) FAS2, complete
Length = 1945
Score = 50.8 bits (120), Expect = 1e-06
Identities = 39/175 (22%), Positives = 74/175 (42%), Gaps = 9/175 (5%)
Frame = +1
Query: 6 GKLAFDIDFHPSDNLVATGLIDGDLHLYRYS-SDNTNSDPV--RLLEIHAHTESCRAARF 62
GK +DFHP +AT D D+ ++ + + PV L + H+ + RF
Sbjct: 37 GKPVLTLDFHPHSATLATAGADFDIKFWQIKPAGSPKKLPVVSYLSNLSYHSSAVNVIRF 216
Query: 63 INGGRALLTGSPDFSIL-----ATDVETGSTIARLDNAHEAAVNRLVNLTEST-VASGDD 116
+ G L +G+ ++ +TD + ++ +H + L T++T + SG
Sbjct: 217 SSSGEXLASGADGGDLIIWKLHSTDAGQTWKVLKMLRSHHKDILDLQWSTDATYIISGSV 396
Query: 117 DGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNK 171
D C +WD + + + + H Y+ + + + + S D T C + NK
Sbjct: 397 DNCCIIWDVNKGTNLQTLDTHAHYVQGVAWDPLGKYVTSLSSDRT---CRIYMNK 552
Score = 31.2 bits (69), Expect = 0.80
Identities = 32/127 (25%), Positives = 57/127 (44%), Gaps = 10/127 (7%)
Frame = +1
Query: 137 HEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVV 196
H I+DIT++SDA L +S DG S+ +++ + SE + V K+GR V
Sbjct: 1012 HYAAITDITWSSDAHYLALSSQDGFCSLVEFENDELGSPYSLSERK----VSNKDGRSTV 1179
Query: 197 ----------CGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGM 246
G+ + ++ +D +D V +S +I ++ + + +T + GM
Sbjct: 1180 QTTNDTVTVPTGNVSAVLAESKKMESEDTADDVVIEASGNIGAVV-TESGKTVTQEKAGM 1356
Query: 247 INLVGIL 253
I G L
Sbjct: 1357 IQSTGNL 1377
>TC209377 similar to UP|Q9FLX9 (Q9FLX9) Notchless protein homolog
(At5g52820), partial (33%)
Length = 669
Score = 50.4 bits (119), Expect = 1e-06
Identities = 31/113 (27%), Positives = 55/113 (48%), Gaps = 1/113 (0%)
Frame = +2
Query: 96 HEAAVNRLVNLTEST-VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLL 154
H+ VN + + VAS D +K+W+ +F H + I++++D+ LL
Sbjct: 122 HQQLVNHVYFSPDGQWVASASFDKSVKLWNGTTGKFVTAFRGHVGPVYQISWSADSRLLL 301
Query: 155 ATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLY 207
+ S D TL V ++R K++ DE+ SV +G KV G + ++ L+
Sbjct: 302 SGSKDSTLKVWDIRTRKLKQDLPGHADEVFSVDWSPDGEKVASGGKDKVLKLW 460
>TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%)
Length = 1048
Score = 49.7 bits (117), Expect = 2e-06
Identities = 38/169 (22%), Positives = 75/169 (43%), Gaps = 1/169 (0%)
Frame = +2
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+D+ F P + A+ D ++ S D P+R++ H C ++ +
Sbjct: 125 WDVQFSPVGHYFASSSHDRTARIW--SMDRIQ--PLRIMAGHLSDVDC--VQWHANCNYI 286
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-VASGDDDGCIKVWDTRER 128
TGS D ++ DV++G + R+ H + L + +ASGD+DG I +WD
Sbjct: 287 ATGSSDKTVRLWDVQSGECV-RVFVGHRGMILSLAMSPDGRYMASGDEDGTIMMWDLSSG 463
Query: 129 SCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSE 177
C H + + F+S+ + + S D T+ + ++ + +++E
Sbjct: 464 RCLTPLIGHTSCVWSLAFSSEGSVIASGSADCTVKLWDVNTSTKVSRAE 610
Score = 40.4 bits (93), Expect = 0.001
Identities = 44/215 (20%), Positives = 85/215 (39%)
Frame = +2
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
+ S D I++W T+ + ++ H + D+ F+ ++S D T + ++ R
Sbjct: 32 ILSSSADSTIRLWSTKLNANLVCYKGHNYPVWDVQFSPVGHYFASSSHDRTARIWSMDRI 211
Query: 171 KVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTM 230
+ ++ V N + GS + L+ +C FV +
Sbjct: 212 QPLRIMAGHLSDVDCVQWHANCNYIATGSSDKTVRLWDVQS-GECVRVFVGHRGMILSLA 388
Query: 231 LKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSI 290
+ D + +G E+G I + + R + P+ H+ +AFS + + S
Sbjct: 389 MSPDGRYMASGDEDGTIMMWDLSSGRCLTPLIGHTSCVWS-------LAFSSEGSVIASG 547
Query: 291 AHDQMLKLWDLDNILQGSRNTQRNENGGVDNDVDS 325
+ D +KLWD++ + SR E GG N + S
Sbjct: 548 SADCTVKLWDVNTSTKVSR---AEEKGGSANRLRS 643
>TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (57%)
Length = 1124
Score = 47.8 bits (112), Expect = 8e-06
Identities = 33/162 (20%), Positives = 70/162 (42%), Gaps = 4/162 (2%)
Frame = +1
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTV 111
H+E + + + + S D ++ D G ++ H+ AV + N S +
Sbjct: 259 HSEGISDLAWSSDSHYICSASDDRTLRIWDATVGGGCIKILRGHDDAVFCVNFNPQSSYI 438
Query: 112 ASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNK 171
SG D IKVWD + C ++ + H ++ + + D +++ S DG+ + +
Sbjct: 439 VSGSFDETIKVWDVKTGKCVHTIKGHTMPVTSVHYNRDGNLIISASHDGSCKIWDTETGN 618
Query: 172 VQAQSEFSEDELLSVVLMK---NGRKVVCGSQTGIMLLYSWG 210
+ ED+ +V K NG+ ++ + + L+++G
Sbjct: 619 L--LKTLIEDKAPAVSFAKFSPNGKLILAATLNDTLKLWNYG 738
Score = 36.6 bits (83), Expect = 0.019
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Frame = +1
Query: 96 HEAAVNRLVNLTESTV-ASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLL 154
HE AV+ + + T+ AS D + +W + + C+ H + ISD+ ++SD+ +
Sbjct: 133 HENAVSCVKFSNDGTLLASASLDKTLIIWSSATLTLCHRLVGHSEGISDLAWSSDSHYIC 312
Query: 155 ATSGDGTLSV 164
+ S D TL +
Sbjct: 313 SASDDRTLRI 342
Score = 29.6 bits (65), Expect = 2.3
Identities = 14/56 (25%), Positives = 27/56 (48%), Gaps = 2/56 (3%)
Frame = +2
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLAT--SGDGTLSV 164
+ G +D C+ +WD +++ E H D + +T K+ + +GD T+ V
Sbjct: 827 IVGGSEDHCVYIWDLQQK-LVQKLEGHTDTVISVTCHPTENKIASAGLAGDRTVRV 991
Score = 28.9 bits (63), Expect = 4.0
Identities = 19/92 (20%), Positives = 38/92 (40%)
Frame = +1
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
F ++F+P + + +G D + ++ + + I HT + + G +
Sbjct: 403 FCVNFNPQSSYIVSGSFDETIKVWDVKTGKC------VHTIKGHTMPVTSVHYNRDGNLI 564
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVN 101
++ S D S D ETG+ + L AV+
Sbjct: 565 ISASHDGSCKIWDTETGNLLKTLIEDKAPAVS 660
>TC216518 weakly similar to UP|Q9LW87 (Q9LW87) Coatomer protein complex, beta
prime; beta'-COP protein, partial (29%)
Length = 1189
Score = 47.0 bits (110), Expect = 1e-05
Identities = 24/107 (22%), Positives = 48/107 (44%)
Frame = +1
Query: 100 VNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGD 159
V+ + + + SG DD KVWD R+C + E HE+ ++ I + ++ S D
Sbjct: 622 VDYFITNDKQYLLSGSDDYTAKVWDYHSRNCVQTLEGHENNVTAICAHPELPIIITASED 801
Query: 160 GTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLL 206
T+ + + ++Q F + S+ K ++ G G +++
Sbjct: 802 STVKIWDAVTYRLQTTLNFGLKRVWSIGYKKGSSQLAFGCDQGFLIV 942
Score = 30.8 bits (68), Expect = 1.0
Identities = 36/165 (21%), Positives = 65/165 (38%), Gaps = 5/165 (3%)
Frame = +1
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
F +N + D ++H+Y Y +++E H + R+ +++ S
Sbjct: 244 FIARENWIVAATDDKNIHVYNYDKME------KIVEFAEHKDYIRSLAVHPVLPYVISAS 405
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTE--STVASGDDDGCIKVWDTRERSCC 131
D + + G + H V ++ + ST AS DG +K+W +
Sbjct: 406 DDQVLKLWNWRKGWSCYENFEGHSHYVMQVAFNPKDPSTFASASLDGTLKIWSLDSSAPN 585
Query: 132 NSFEAHEDYIS--DITFASDAMKLLATSGDGTLSVCNLR-RNKVQ 173
+ E H+ ++ D +D LL+ S D T V + RN VQ
Sbjct: 586 FTLEGHQKGVNCVDYFITNDKQYLLSGSDDYTAKVWDYHSRNCVQ 720
>TC207353 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (58%)
Length = 898
Score = 47.0 bits (110), Expect = 1e-05
Identities = 35/172 (20%), Positives = 77/172 (44%), Gaps = 10/172 (5%)
Frame = +3
Query: 105 NLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
N S + SG D IKVWD + C ++ + H ++ + + D +++ S DG+ +
Sbjct: 21 NPQSSYIVSGSFDETIKVWDVKTGKCVHTIKGHTMPVTSVHYNRDGTLIISASHDGSCKI 200
Query: 165 CNLRRNKVQAQSEFSEDELLSVVLMK---NGRKVVCGSQTGIMLLYSWGCFKDCSDRFVD 221
+ + ED+ +V K NG+ ++ + + L+++G S +F+
Sbjct: 201 WDTGTGNL--LKTLIEDKAPAVSFAKFSPNGKFILAATLNDTLKLWNYG-----SGKFLK 359
Query: 222 LSSNSID-------TMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSE 266
+ S ++ T + I++GSE+ + + + +IQ + H++
Sbjct: 360 IYSGHVNRVYCITSTFSVTNGRYIVSGSEDRCVYIWDLQAKNMIQKLEGHTD 515
Score = 36.2 bits (82), Expect = 0.025
Identities = 24/114 (21%), Positives = 51/114 (44%), Gaps = 9/114 (7%)
Frame = +3
Query: 60 ARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-------VA 112
A+F G+ +L + + ++ + +G + + VNR+ +T + +
Sbjct: 267 AKFSPNGKFILAATLNDTLKLWNYGSGKFL----KIYSGHVNRVYCITSTFSVTNGRYIV 434
Query: 113 SGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLAT--SGDGTLSV 164
SG +D C+ +WD + ++ E H D + +T K+ + +GD T+ V
Sbjct: 435 SGSEDRCVYIWDLQAKNMIQKLEGHTDTVISVTCHPTENKIASAGLAGDRTVRV 596
>TC206580 weakly similar to UP|Q9JKK6 (Q9JKK6) G beta-like protein GBL,
partial (32%)
Length = 997
Score = 45.8 bits (107), Expect = 3e-05
Identities = 36/144 (25%), Positives = 65/144 (45%), Gaps = 2/144 (1%)
Frame = +3
Query: 28 GDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGS 87
G+ H+ + ++ + PV + +HT + A F G + +GS D ++ D+
Sbjct: 375 GNPHIRLFDVNSNSPQPV--MSYDSHTNNVMAVGFQCDGNWMYSGSEDGTVKIWDLRAPG 548
Query: 88 TIARLDNAHEAAVNRLV-NLTESTVASGDDDGCIKVWDTRERSC-CNSFEAHEDYISDIT 145
++ AAVN +V + ++ + SGD +G I+VWD SC C + + +T
Sbjct: 549 CQREYES--RAAVNTVVLHPNQTELISGDQNGNIRVWDLTANSCSCELVPEVDTAVRSLT 722
Query: 146 FASDAMKLLATSGDGTLSVCNLRR 169
D ++A + T V L R
Sbjct: 723 VMWDGSLVVAANNHRTCYVWRLLR 794
Score = 36.2 bits (82), Expect = 0.025
Identities = 21/88 (23%), Positives = 42/88 (46%), Gaps = 1/88 (1%)
Frame = +3
Query: 113 SGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKV 172
SG +DG +K+WD R C +E+ ++ + + +L++ +G + V +L N
Sbjct: 498 SGSEDGTVKIWDLRAPGCQREYESRA-AVNTVVLHPNQTELISGDQNGNIRVWDLTANSC 674
Query: 173 QAQSEFSEDELL-SVVLMKNGRKVVCGS 199
+ D + S+ +M +G VV +
Sbjct: 675 SCELVPEVDTAVRSLTVMWDGSLVVAAN 758
>TC222867 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Fragment),
partial (23%)
Length = 591
Score = 45.1 bits (105), Expect = 5e-05
Identities = 28/123 (22%), Positives = 54/123 (43%), Gaps = 1/123 (0%)
Frame = +1
Query: 43 DPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNR 102
D V ++ H + F + ++T S D +I + GS + + H ++V R
Sbjct: 16 DLVSVVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAISDGSCLKTFEG-HTSSVLR 192
Query: 103 LVNLTEST-VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGT 161
+ +T T + S DG +K+W + C +++ HED + + KL GD
Sbjct: 193 ALFVTRGTQIVSCGADGLVKLWTVKTNECVATYDHHEDKVWALAVGRKTEKLATGGGDAV 372
Query: 162 LSV 164
+++
Sbjct: 373 VNL 381
Score = 43.9 bits (102), Expect = 1e-04
Identities = 19/100 (19%), Positives = 47/100 (47%)
Frame = +1
Query: 108 ESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNL 167
+ V + D I++W + SC +FE H + F + ++++ DG + + +
Sbjct: 85 DQCVVTASGDKTIRIWAISDGSCLKTFEGHTSSVLRALFVTRGTQIVSCGADGLVKLWTV 264
Query: 168 RRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLY 207
+ N+ A + ED++ ++ + + K+ G ++ L+
Sbjct: 265 KTNECVATYDHHEDKVWALAVGRKTEKLATGGGDAVVNLW 384
>TC208434 similar to PIR|T01391|T01391 WD-repeat protein T4I9.10 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(68%)
Length = 1364
Score = 45.1 bits (105), Expect = 5e-05
Identities = 34/117 (29%), Positives = 53/117 (45%), Gaps = 6/117 (5%)
Frame = +1
Query: 59 AARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES--TVASGDD 116
+ +F GR L+ G+ D SI D+ R+ AH++ VN + ES + SG D
Sbjct: 415 SVKFSTDGRELVAGTSDCSICVYDLGADKLSLRIP-AHQSDVNTVCFADESGHLIYSGSD 591
Query: 117 DGCIKVWDTR----ERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRR 169
D IKVWD R + H + I+ I D L++ D T + ++R+
Sbjct: 592 DSFIKVWDRRCFVAKGQPAGILMGHLEGITFIDSRGDGRYLISNGKDQTTKLWDIRK 762
>TC209542
Length = 1445
Score = 43.1 bits (100), Expect = 2e-04
Identities = 36/152 (23%), Positives = 64/152 (41%), Gaps = 11/152 (7%)
Frame = +2
Query: 24 GLIDG-----DLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSI 78
G +DG D+ + R +SDN PV + + N G +L G D ++
Sbjct: 476 GSVDGTVRTFDIRIGRETSDNLGQ-PVNCVSMS------------NDGNCILAGCLDSTL 616
Query: 79 LATDVETGSTIARLDNAHEAAVNRLVNL------TESTVASGDDDGCIKVWDTRERSCCN 132
D TG + ++ N+ L T++ V +DG I WD + S +
Sbjct: 617 RLLDRSTGELLQE----YKGHTNKSYKLDCCLTNTDAHVTGVSEDGFIYFWDLVDASVVS 784
Query: 133 SFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
F+AH ++ +++ ++ +S DGT+ V
Sbjct: 785 RFKAHTSVVTSVSYHPKENCMVTSSVDGTIRV 880
>TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nuclear
ribonucleoprotein Prp4 (U4/U6 snRNP 60 kDa protein) (WD
splicing factor Prp4) (hPrp4), partial (34%)
Length = 1206
Score = 42.4 bits (98), Expect = 3e-04
Identities = 39/164 (23%), Positives = 63/164 (37%), Gaps = 1/164 (0%)
Frame = +1
Query: 9 AFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRA 68
A D+ + P + +AT D + S L H + F G+
Sbjct: 208 ATDVAYSPVHDHLATASADRTAKYWNQGS--------LLKTFEGHLDRLARIAFHPSGKY 363
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDD-DGCIKVWDTRE 127
L T S D + D+ETG + L H +V L + ++A+ D +VWD R
Sbjct: 364 LGTASFDKTWRLWDIETGDELL-LQEGHSRSVYGLAFHNDGSLAASCGLDSLARVWDLRT 540
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNK 171
+ E H + I+F+ + L D T + +LR+ K
Sbjct: 541 GRSILALEGHVKPVLGISFSPNGYHLATGGEDNTCRIWDLRKKK 672
Score = 28.5 bits (62), Expect = 5.2
Identities = 29/167 (17%), Positives = 61/167 (36%), Gaps = 2/167 (1%)
Frame = +1
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + FH +L A+ +D ++ + + +L + H + F G L
Sbjct: 457 YGLAFHNDGSLAASCGLDSLARVWDLRTGRS------ILALEGHVKPVLGISFSPNGYHL 618
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST--VASGDDDGCIKVWDTRE 127
TG D + D+ + + AH ++++ + + D KVW R+
Sbjct: 619 ATGGEDNTCRIWDLRKKKSFYTIP-AHSNLISQVKFEPHEGYFLVTASYDMTAKVWSGRD 795
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQA 174
+ HE ++ + D ++ S D T+ + + QA
Sbjct: 796 FKPVKTLSGHEAKVTSVDVLGDGGSIVTVSHDRTIKLWSSNPTDEQA 936
>BI787980 similar to SP|P25387|GBLP Guanine nucleotide-binding protein beta
subunit-like protein. {Chlamydomonas reinhardtii},
partial (33%)
Length = 421
Score = 40.4 bits (93), Expect = 0.001
Identities = 32/126 (25%), Positives = 54/126 (42%), Gaps = 7/126 (5%)
Frame = +3
Query: 90 ARLDNAHEAAVNRLVNLTEST--VASGDDDGCIKVW----DTRERSCCNS-FEAHEDYIS 142
A LD + AV L E+ + + D + VW D E + H Y+
Sbjct: 36 AELDGHNGKAVTALATTRENPNLLLTASRDKSLLVWQLSNDGEEYGFARRRLQGHSHYVE 215
Query: 143 DITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTG 202
D+ +SD L+ S DGTL + +L + ++LSV + R++V GS+
Sbjct: 216 DVVISSDGQFALSGSWDGTLRLWDLNTGITTRRFVGHTKDVLSVAFSADNRQIVSGSRDK 395
Query: 203 IMLLYS 208
+ L++
Sbjct: 396 TVKLWN 413
>TC207267 similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (53%)
Length = 1254
Score = 40.4 bits (93), Expect = 0.001
Identities = 49/180 (27%), Positives = 76/180 (42%), Gaps = 11/180 (6%)
Frame = +2
Query: 10 FDIDFHPSDNLVATGLIDGDLH--LYRYSSDNTNSDPVRLLEIHAHTESCRAARFI-NGG 66
+ ID+ P LV L GD + +Y + + + V HT S ++
Sbjct: 224 YAIDWSP---LVPGKLASGDCNNCIYLWEPTSAGTWNVDNAPFIGHTASVEDLQWSPTES 394
Query: 67 RALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLV--NLTESTVASGDDDGCIKVWD 124
+ S D +I D G + A AH A VN + L +ASG DDG I + D
Sbjct: 395 HVFASCSVDGNIAIWDTRLGKSPAASFKAHNADVNVMSWNRLASCMLASGSDDGTISIRD 574
Query: 125 TR----ERSCCNSFEAHEDYISDITFA-SDAMKLLATSGDGTLSVCNLRRNK-VQAQSEF 178
R S FE H+ I+ I ++ +A L +S D L++ +L K + ++EF
Sbjct: 575 LRLLKEGDSVVAHFEYHKHPITSIEWSPHEASSLAVSSSDNQLTIWDLSLEKDEEEEAEF 754
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,120,693
Number of Sequences: 63676
Number of extensions: 166933
Number of successful extensions: 1135
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 1046
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1116
length of query: 378
length of database: 12,639,632
effective HSP length: 99
effective length of query: 279
effective length of database: 6,335,708
effective search space: 1767662532
effective search space used: 1767662532
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142222.5


