

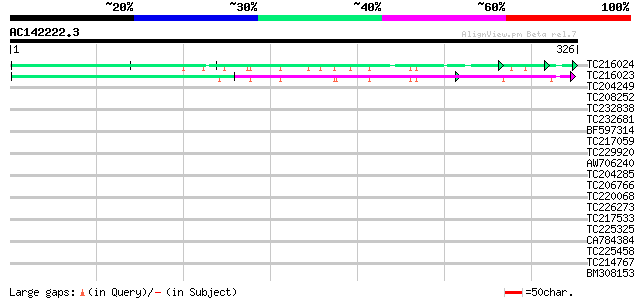
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.3 + phase: 0
(326 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216024 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F2... 56 3e-08
TC216023 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F2... 50 1e-06
TC204249 UP|Q9SB11 (Q9SB11) Glycinin, complete 40 0.002
TC208252 weakly similar to UP|Q9HCC8 (Q9HCC8) Osteoblast differe... 39 0.002
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 36 0.027
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 35 0.060
BF597314 similar to PIR|S20007|S20 beta-conglycinin alpha chain ... 34 0.10
TC217059 similar to UP|Q8L7W3 (Q8L7W3) At2g29210/F16P2.41, parti... 33 0.13
TC229920 weakly similar to UP|O64855 (O64855) Expressed protein ... 33 0.17
AW706240 similar to GP|21593823|gb| 40S ribosomal protein S17-3 ... 33 0.23
TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 33 0.23
TC206766 similar to PIR|T47576|T47576 FKBP12 interacting protein... 33 0.23
TC220068 similar to GB|AAL31246.1|16974485|AY061919 AT4g32480/F8... 32 0.39
TC226273 similar to GB|AAP37745.1|30725446|BT008386 At5g61156 {A... 32 0.51
TC217533 similar to UP|Q9LUS2 (Q9LUS2) Chloroplast outer envelop... 32 0.51
TC225325 similar to UP|R17D_ARATH (Q9LZ17) 40S ribosomal protein... 32 0.51
CA784384 31 0.66
TC225458 UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37, complete 31 0.66
TC214767 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein... 31 0.66
BM308153 similar to GP|6960465|gb|A ORF4 hoar {Spodoptera exigua... 31 0.66
>TC216024 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F23H14.7
{Arabidopsis thaliana;} , partial (14%)
Length = 1433
Score = 55.8 bits (133), Expect = 3e-08
Identities = 60/263 (22%), Positives = 107/263 (39%), Gaps = 22/263 (8%)
Frame = +2
Query: 70 DDYSLDEIFPNRRQKHQAHKRHYHYDDED----SLDSLLPKQHQAERGHTRYDD-EDSLD 124
D+ S DE ++ H+ H++H HYD D S K ++ + YDD E S +
Sbjct: 365 DNDSDDESRKTSKRSHRKHRKHSHYDSGDHEKRKEKSSKRKTKKSSSETSDYDDFESSSE 544
Query: 125 EVLPKQRQAKR-----GRSHYDDDDSL-DEVLPKQHHAKRGHPHYDDDDDLP----DGVL 174
E K++Q+K+ RS D DS+ DE+ ++ H KR P D D ++
Sbjct: 545 EERRKKKQSKKLRDRGSRSDSSDSDSIADELSKRKRHHKRQRPSKFSGSDYSTEEGDSLV 724
Query: 175 PKQ---QAKKRHPHYDNDEDSL-HEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKR 230
K+ + KRH ++ E L +D P+ ++ + H+ N+ + ++
Sbjct: 725 RKRNHGKHSKRHRRSESSESDLSSDDAPRKKGHRKHQKHHKRSHNV-------ELRSSDS 883
Query: 231 HPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLH 290
H+ + S + L +N + E + + D K H D++ + +
Sbjct: 884 DYHHSHGQRSRSKSLEENSEDESKRSMHKKSDRHHHHHRHHHHHHHKHRHHLDDERNHMQ 1063
Query: 291 EDLPK---NHQAKRGRPLYDDTD 310
+ PK H K + D D
Sbjct: 1064QHSPKFNGKHDEKLDKTETDKKD 1132
Score = 53.9 bits (128), Expect = 1e-07
Identities = 69/311 (22%), Positives = 122/311 (39%), Gaps = 28/311 (9%)
Frame = +2
Query: 2 KALERQSAGAFTSVSQARRARQSAGAFTSVSQARRGHPRYDDDHSLDEFLADRRRKKHLA 61
K +ER+ A A + +R D D+ D+ ++ H
Sbjct: 239 KIMERKEAPLKWEQKLEAAAETKERKLKATKHKKRSGSDSDSDNDSDDESRKTSKRSHRK 418
Query: 62 LRRHPHYDD-DYSLDEIFPNRRQKHQAHKRHYHYDD-EDSLDSLLPKQHQA----ERGHT 115
R+H HYD D+ + ++R+ ++ YDD E S + K+ Q+ +RG +
Sbjct: 419 HRKHSHYDSGDHEKRKEKSSKRKTKKSSSETSDYDDFESSSEEERRKKKQSKKLRDRG-S 595
Query: 116 RYDDEDS---LDEVLPKQRQAKRGR------SHYDDDDSLDEVLPKQH--HAKRGHPHYD 164
R D DS DE+ ++R KR R S Y ++ V + H H+KR
Sbjct: 596 RSDSSDSDSIADELSKRKRHHKRQRPSKFSGSDYSTEEGDSLVRKRNHGKHSKRHRRSES 775
Query: 165 DDDDLPDGVLPKQQAKKRHPHYDNDEDSLH---EDLPKNHQAKRGRSHYDVEDNLLDGVL 221
+ DL P+++ ++H + ++ D +H + +E+N D
Sbjct: 776 SESDLSSDDAPRKKGHRKHQKHHKRSHNVELRSSDSDYHHSHGQRSRSKSLEENSEDE-- 949
Query: 222 PKQPQAKK--------RHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQR 273
K+ KK RH H+ + H D +NH ++ P+++ D L K
Sbjct: 950 SKRSMHKKSDRHHHHHRHHHHHHHKHRHHLDDERNHM-QQHSPKFNGKH---DEKLDKTE 1117
Query: 274 QAKKLHPHYDN 284
KK H+++
Sbjct: 1118TDKKDGGHHES 1150
Score = 50.1 bits (118), Expect = 1e-06
Identities = 53/227 (23%), Positives = 89/227 (38%), Gaps = 20/227 (8%)
Frame = +2
Query: 120 EDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDD-DDDLPDGVLPKQQ 178
E L K+R S D DD + + H R H HYD D + K++
Sbjct: 308 ERKLKATKHKKRSGSDSDSDNDSDDESRKTSKRSHRKHRKHSHYDSGDHEKRKEKSSKRK 487
Query: 179 AKKRHPH---YDNDEDSLHEDLPKNHQAKR-----GRSHYDVEDNLLDGVLPKQPQAKKR 230
KK YD+ E S E+ K Q+K+ RS D++ D + ++ K++
Sbjct: 488 TKKSSSETSDYDDFESSSEEERRKKKQSKKLRDRGSRSDSSDSDSIADELSKRKRHHKRQ 667
Query: 231 HP-------HYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYD 283
P + +GDSL ++R + + DL P+++ +K H+
Sbjct: 668 RPSKFSGSDYSTEEGDSLVRKRNHGKHSKRHRRSESSESDLSSDDAPRKKGHRKHQKHHK 847
Query: 284 NDED----SLHEDLPKNHQAKRGRPLYDDTDSLDEVLPARRRRHYRS 326
+ S D +H +R R + +S DE ++R H +S
Sbjct: 848 RSHNVELRSSDSDYHHSH-GQRSRSKSLEENSEDE---SKRSMHKKS 976
>TC216023 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F23H14.7
{Arabidopsis thaliana;} , partial (15%)
Length = 1437
Score = 50.4 bits (119), Expect = 1e-06
Identities = 62/287 (21%), Positives = 111/287 (38%), Gaps = 29/287 (10%)
Frame = +3
Query: 2 KALERQSAGAFTSVSQARRARQSAGAFTSVSQARRGHPRYDDDHSLDEFLADRRRKKHLA 61
K +ER+ A A +R D D+ D+ ++ H
Sbjct: 228 KIMERKEAPLKWEQKLEAAAEAKERKLKVAKHTKRSGSDSDSDYDSDDESRKTSKRSHRK 407
Query: 62 LRRHPHYDD-DYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDD- 119
R+H HYD D+ + ++R+ + Y +DS S ++ + ++ + D
Sbjct: 408 HRKHSHYDSGDHEKRKEKSSKRKTKKWSSESSDYSSDDSESSFEEERRRKKKQSKKLRDR 587
Query: 120 ---------EDSLDEVLPKQRQAKRGR------SHYDDDDSLDEVLPKQH--HAKRGHPH 162
+ + DE+ ++RQ KR R S Y D+ V + H H+KR
Sbjct: 588 GSRSDSSNYDSTADELSKRKRQHKRQRPSKFSGSDYSTDEGDSLVQKRNHGMHSKRHRQS 767
Query: 163 YDDDDDLPDGVLP-KQQAKKRHPHY---DNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLD 218
+ DL P K+ +K H H+ N E L + ++H +R RS +E+N D
Sbjct: 768 ESSESDLSSDDAPHKKGHRKHHKHHKRSHNVELRLSDSDYRSH-GQRSRSK-SLEENSED 941
Query: 219 GVLPKQPQAKK------RHPHYDNDGDSLHEDLPKNHQAERGQPRYD 259
+ K RH H+ + H D +N+ ++ P+++
Sbjct: 942 ESKKRSMDKKSGRHHHHRHHHHRHHHHRHHLDDERNYM-QQHSPKFN 1079
Score = 46.2 bits (108), Expect = 2e-05
Identities = 53/225 (23%), Positives = 93/225 (40%), Gaps = 29/225 (12%)
Frame = +3
Query: 130 QRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDD-DDDLPDGVLPKQQAKKRHPH--- 185
+R S YD DD + + H R H HYD D + K++ KK
Sbjct: 327 KRSGSDSDSDYDSDDESRKTSKRSHRKHRKHSHYDSGDHEKRKEKSSKRKTKKWSSESSD 506
Query: 186 --YDNDEDSLHED-LPKNHQAKR-----GRSHYDVEDNLLDGVLPKQPQAKKRHP----- 232
D+ E S E+ K Q+K+ RS D+ D + ++ Q K++ P
Sbjct: 507 YSSDDSESSFEEERRRKKKQSKKLRDRGSRSDSSNYDSTADELSKRKRQHKRQRPSKFSG 686
Query: 233 --HYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHY---DNDED 287
+ ++GDSL + ++R + + DL P ++ +K H H+ N E
Sbjct: 687 SDYSTDEGDSLVQKRNHGMHSKRHRQSESSESDLSSDDAPHKKGHRKHHKHHKRSHNVEL 866
Query: 288 SLHEDLPKNH-QAKRGRPLYDDTD------SLDEVLPARRRRHYR 325
L + ++H Q R + L ++++ S+D+ + R H+R
Sbjct: 867 RLSDSDYRSHGQRSRSKSLEENSEDESKKRSMDK--KSGRHHHHR 995
>TC204249 UP|Q9SB11 (Q9SB11) Glycinin, complete
Length = 1916
Score = 39.7 bits (91), Expect = 0.002
Identities = 41/167 (24%), Positives = 67/167 (39%), Gaps = 12/167 (7%)
Frame = +1
Query: 120 EDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLP-----KQHHAKRGHPH-YDDDDDLPDGV 173
E L + PK Q ++ +D+D DE +P + H KR D+D+D P
Sbjct: 838 EGGLSVISPKW-QEQQDEDEDEDEDDEDEQIPSHPPRRPSHGKREQDEDEDEDEDKPRPS 1014
Query: 174 LPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPH 233
P Q +++ D DED ED P+ + R + P+Q + ++R
Sbjct: 1015RPSQGKREQDQDQDEDEDE-DEDQPRKSREWRSKKTQPRR--------PRQEEPRERGCE 1167
Query: 234 YDNDGD------SLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQ 274
N + LHE++ + +A+ P+ L LP RQ
Sbjct: 1168TRNGVEENICTLKLHENIARPSRADFYNPKAGRISTLNSLTLPALRQ 1308
Score = 30.4 bits (67), Expect = 1.1
Identities = 38/172 (22%), Positives = 70/172 (40%), Gaps = 11/172 (6%)
Frame = +1
Query: 166 DDDLPDGVLPKQQAK-----KRHPHYDNDED---SLHEDLPKNHQAKRGRSHYDVEDNLL 217
D + P+ + +QQ K K+ H +E+ S+ K+ A+ ++ D+ + L
Sbjct: 622 DIEYPETMQQQQQQKSHGGRKQGQHQQEEEEEGGSVLSGFSKHFLAQSFNTNEDIAEKL- 798
Query: 218 DGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKK 277
+ P +++ G S+ + Q E DD+D+ P++ K
Sbjct: 799 -----QSPDDERKQIVTVEGGLSVISPKWQEQQDEDEDEDEDDEDEQIPSHPPRRPSHGK 963
Query: 278 LHPHYDNDEDSLHEDLPKNHQAKRGRPLYD---DTDSLDEVLPARRRRHYRS 326
D DED ED P+ + +G+ D D D ++ R+ R +RS
Sbjct: 964 REQDEDEDED---EDKPRPSRPSQGKREQDQDQDEDEDEDEDQPRKSREWRS 1110
>TC208252 weakly similar to UP|Q9HCC8 (Q9HCC8) Osteoblast differentiation
promoting factor (AESP1935), partial (4%)
Length = 635
Score = 39.3 bits (90), Expect = 0.002
Identities = 40/176 (22%), Positives = 62/176 (34%), Gaps = 9/176 (5%)
Frame = +3
Query: 36 RGHPRYDDDHSLDEFLADRR---------RKKHLALRRHPHYDDDYSLDEIFPNRRQKHQ 86
+G D+D S +E + +R RK + R Y SL+E +R +HQ
Sbjct: 21 KGTTSQDNDSSSEEERSQKRSNREKYDESRKDKTPVTRGRDYSSSSSLEEERSRKRSRHQ 200
Query: 87 AHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSL 146
HKRH + G D+D D + + ++G S
Sbjct: 201 RHKRH-------------------DAGSHSSSDDDRRDRHSSRSKGRRKGSSR------- 302
Query: 147 DEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQ 202
E + H+KR H H K ++ R Y N++DS H K +
Sbjct: 303 -EKSRSEKHSKR-HKH------------RKHESPNRSSRYSNEKDSSHSRKEKRRR 428
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 35.8 bits (81), Expect = 0.027
Identities = 39/169 (23%), Positives = 65/169 (38%), Gaps = 13/169 (7%)
Frame = +2
Query: 118 DDED--SLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVL- 174
DDE+ S+DEV+ + DDDD D+ ++ A G DD+DD +G +
Sbjct: 287 DDEEYESVDEVVD-DAEDDEDEEEDDDDDEGDDNDDEEDDAPDGGDDDDDEDDEEEGDVQ 463
Query: 175 ----PKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHY------DVEDNLLDGVLPKQ 224
P D+DED ED + + + + Y E+ +
Sbjct: 464 RGGEPDDDDNDSDDDSDDDEDEDEEDEEEQGEEEDLGTEYLIRPLETAEEEEASSDFEPE 643
Query: 225 PQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQR 273
++ D++ E PK ++++ DDDD D P +R
Sbjct: 644 ENGEEEEEDVDDEDCEKAEAPPKRKRSDKDDS--DDDDGGEDDERPSKR 784
Score = 32.0 bits (71), Expect = 0.39
Identities = 43/180 (23%), Positives = 70/180 (38%), Gaps = 4/180 (2%)
Frame = +2
Query: 69 DDDY-SLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVL 127
D++Y S+DE+ + + DD+D D ++ A G DDED +E
Sbjct: 290 DEEYESVDEVVDDAEDDEDEEED----DDDDEGDDNDDEEDDAPDGGDDDDDEDDEEE-- 451
Query: 128 PKQRQAKRGRSHYDDD-DSLDEVLPKQHHAKRGHPHYDDDDDLPDGVL--PKQQAKKRHP 184
+RG DDD DS D+ + + +++DL L P + A++
Sbjct: 452 ---GDVQRGGEPDDDDNDSDDDSDDDEDEDEEDEEEQGEEEDLGTEYLIRPLETAEEEEA 622
Query: 185 HYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHED 244
D + + E+ DV+D D + P +KR D+D D ED
Sbjct: 623 SSDFEPEENGEE-----------EEEDVDDE--DCEKAEAPPKRKRSDKDDSDDDDGGED 763
Score = 30.0 bits (66), Expect = 1.5
Identities = 19/72 (26%), Positives = 29/72 (39%)
Frame = +2
Query: 243 EDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKRG 302
ED ++ + G D++DD PDG D+D +D + + G
Sbjct: 341 EDEEEDDDDDEGDDNDDEEDDAPDG----------------GDDDDDEDDEEEGDVQRGG 472
Query: 303 RPLYDDTDSLDE 314
P DD DS D+
Sbjct: 473 EPDDDDNDSDDD 508
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 34.7 bits (78), Expect = 0.060
Identities = 35/151 (23%), Positives = 62/151 (40%), Gaps = 7/151 (4%)
Frame = +1
Query: 134 KRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSL 193
+R S DDDD+ DE + A G DD+D+ + + + + D+DED
Sbjct: 193 RRRLSFDDDDDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDED 372
Query: 194 HEDLPKNHQAKRGRSHYDV-------EDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLP 246
ED + + + Y + E+ P++ ++ D DG+ E P
Sbjct: 373 EEDEEEQGEEADLGTEYLIRPLVTAEEEEASSDFEPEENGEEEEEDVDDEDGEK-SEAPP 549
Query: 247 KNHQAERGQPRYDDDDDLPDGVLPKQRQAKK 277
K ++++ DD DD G +R +K+
Sbjct: 550 KRKRSDK-----DDSDDDDGGEDDDERPSKR 627
Score = 34.3 bits (77), Expect = 0.078
Identities = 36/139 (25%), Positives = 55/139 (38%), Gaps = 12/139 (8%)
Frame = +1
Query: 118 DDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQ 177
DD+D+ DE + A G DDDD DE ++ +RG DDD+D D ++
Sbjct: 214 DDDDNEDEEDDDEDDAPDGG---DDDDDEDE--EEESDVQRGGEPDDDDNDDDDEDEDEE 378
Query: 178 QAKKR------------HPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQP 225
+++ P +E+ D + DV+D DG + P
Sbjct: 379 DEEEQGEEADLGTEYLIRPLVTAEEEEASSDFEPEENGE--EEEEDVDDE--DGEKSEAP 546
Query: 226 QAKKRHPHYDNDGDSLHED 244
+KR D+D D ED
Sbjct: 547 PKRKRSDKDDSDDDDGGED 603
Score = 27.3 bits (59), Expect = 9.6
Identities = 21/87 (24%), Positives = 34/87 (38%)
Frame = +1
Query: 228 KKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDED 287
++R +D+D D+ E+ DD+DD PDG D+D
Sbjct: 190 RRRRLSFDDDDDNEDEE-------------DDDEDDAPDG----------------GDDD 282
Query: 288 SLHEDLPKNHQAKRGRPLYDDTDSLDE 314
++ ++ + G P DD D DE
Sbjct: 283 DDEDEEEESDVQRGGEPDDDDNDDDDE 363
>BF597314 similar to PIR|S20007|S20 beta-conglycinin alpha chain precursor -
soybean, partial (30%)
Length = 607
Score = 33.9 bits (76), Expect = 0.10
Identities = 27/111 (24%), Positives = 47/111 (42%), Gaps = 12/111 (10%)
Frame = +2
Query: 225 PQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDL---PDGVLPKQRQAKKLHPH 281
PQ +R PH + D +D P++ R QPR D++ + D ++ + +
Sbjct: 236 PQHPEREPHQPGEKDEDEDDAPRSIPFPRPQPRQDEEHEQREEHDWPHEEETRGDNVCED 415
Query: 282 YDNDEDSLHEDL------PKNHQAKRGRPLYDDTDS---LDEVLPARRRRH 323
D DED ++L P +H+ +DD + D+ + RRH
Sbjct: 416 DDEDEDDEQDELQYPFPRPPHHKE*LKHDEHDDDEHHR*SDQSYDSDLRRH 568
Score = 30.4 bits (67), Expect = 1.1
Identities = 26/119 (21%), Positives = 49/119 (40%), Gaps = 3/119 (2%)
Frame = +2
Query: 175 PKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRH--P 232
P Q +R PH ++D +D P++ R + D E + ++ H P
Sbjct: 230 PPPQHPEREPHQPGEKDEDEDDAPRSIPFPRPQPRQDEEH-----------EQREEHDWP 376
Query: 233 HYDND-GDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLH 290
H + GD++ ED ++ E+ + +Y P+ K+ H ++D+D H
Sbjct: 377 HEEETRGDNVCEDDDEDEDDEQDELQYP---------FPRPPHHKE*LKHDEHDDDEHH 526
>TC217059 similar to UP|Q8L7W3 (Q8L7W3) At2g29210/F16P2.41, partial (9%)
Length = 1565
Score = 33.5 bits (75), Expect = 0.13
Identities = 56/316 (17%), Positives = 117/316 (36%), Gaps = 28/316 (8%)
Frame = +1
Query: 15 VSQARRARQSAGAFTSVSQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSL 74
+ + R R S + + +A H Y + + + ++ KH + + Y
Sbjct: 406 LKKQRTQRPSRDSPDTRDEAEETH--YSRESRDPKMNSSQKISKHFPVSKRKGSPAKYRH 579
Query: 75 DEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAK 134
+E P R H A + HY +D + + + +G+ + S+++ + +
Sbjct: 580 EEFSPERVSGHPASEHHYDNNDWSKKGQEIMRDKSSGKGNEFPAQKSSMNK---ETFSRE 750
Query: 135 RGRSHYDDDDSLDEVLPKQH--HAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDS 192
+ + Y D +V + H +AK + H+ + K +++ DS
Sbjct: 751 KPQESYAVDIKKSDVKDQSHSNYAKSSYRHHKSE------TTQDLVGKGDRVNHNASHDS 912
Query: 193 LHEDLPKNHQAKRGRSHY---------DVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHE 243
+ ED K+ + + R + ED D L + +AK+R + L +
Sbjct: 913 VSEDSGKHRREGKDRKRHKRSEKKYTSSDEDYSDDSELEDRKEAKRRR----KEEKKLQK 1080
Query: 244 DLPKNHQAERGQPRYD----------DDDDLPDGVLPKQRQAKKLHPHYDNDED------ 287
+ + + E+ + R + DD+ D +AK++ H + ED
Sbjct: 1081EEKRRRREEKRRRREERRAEKLKMKSKTDDISD-----DEEAKQMGYHQSDSEDEQKKLE 1245
Query: 288 -SLHEDLPKNHQAKRG 302
L ++ +AKRG
Sbjct: 1246IELRNKALESLKAKRG 1293
>TC229920 weakly similar to UP|O64855 (O64855) Expressed protein
(At2g44200/F6E13.34), partial (19%)
Length = 1372
Score = 33.1 bits (74), Expect = 0.17
Identities = 46/229 (20%), Positives = 75/229 (32%), Gaps = 7/229 (3%)
Frame = +3
Query: 80 NRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSH 139
N+ + H+ KR H + L D ED E +R+ K G
Sbjct: 6 NKEKAHKKEKRKKHRSSKSKHKKLS-------------DSEDDTTE----RRKRKTGNED 134
Query: 140 YDDDDSLDEVLPKQHHAKRGHPHYDDDDD---LPDGVLPKQQAKKRHPHYD----NDEDS 192
D ++HH + Y + + Q+ ++R P++ N +D
Sbjct: 135 SD----------RKHHKAQSDSEYQSSEGEMRRRKDRIEDQKYRERSPNHQQRQRNGKDY 284
Query: 193 LHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAE 252
+ +N+ R +S V+ LD K+R HY+ D + + NHQ
Sbjct: 285 KEDAGDRNYN--RSKSERSVQKGQLDSGYESSEGEKRRKNHYE---DKKYRERSPNHQ-- 443
Query: 253 RGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKR 301
QRQ D ED H + NHQ ++
Sbjct: 444 -------------------QRQRNGRDYKEDTGEDKKHRERSPNHQQRQ 533
>AW706240 similar to GP|21593823|gb| 40S ribosomal protein S17-3 {Arabidopsis
thaliana}, partial (52%)
Length = 400
Score = 32.7 bits (73), Expect = 0.23
Identities = 22/99 (22%), Positives = 40/99 (40%), Gaps = 10/99 (10%)
Frame = +1
Query: 124 DEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQA---- 179
D LP +++ R +H+ ++ ++ H + + HP LP + + Q+
Sbjct: 70 DPRLPHKQKTSRRSNHHPFQEATEQDHKFLHTSHKSHP*RSRSQHLPQTPIKESQSPHKL 249
Query: 180 KKRH------PHYDNDEDSLHEDLPKNHQAKRGRSHYDV 212
+ RH PH D +H P+N R H D+
Sbjct: 250 RPRHLRHQHRPHRSQQRDPVHTSFPRNQ--XHPRHHTDI 360
>TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (32%)
Length = 1539
Score = 32.7 bits (73), Expect = 0.23
Identities = 30/112 (26%), Positives = 44/112 (38%), Gaps = 3/112 (2%)
Frame = +2
Query: 54 RRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKR---HYHYDDEDSLDSLLPKQHQA 110
R H L H H + I N R+ HQAH+R H+H + + L L + HQ
Sbjct: 473 RNHHAHQRLSLHIHQNHQRLSLLIHQNHRRNHQAHQRLCLHFHQNHQRIL-LLFHQNHQR 649
Query: 111 ERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPH 162
H + SL + + + R+H+ S L Q + R H H
Sbjct: 650 SHQH----HQRSLLQHRQRSHVHQSSRNHFHQSLSCHHFL--QFQSTRSHFH 787
Score = 28.5 bits (62), Expect = 4.3
Identities = 37/162 (22%), Positives = 61/162 (36%), Gaps = 5/162 (3%)
Frame = +2
Query: 54 RRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKR---HYHYDDEDSLDSLLPKQHQA 110
R + H L H H + I N R+ H AH+R H H + L L+ + H+
Sbjct: 386 RNHQAHQRLSLHIHQNHQRLSLLIHQNHRRNHHAHQRLSLHIH-QNHQRLSLLIHQNHR- 559
Query: 111 ERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLP 170
R H + +R H+ + +L Q+H +R H H+
Sbjct: 560 -RNHQAH----------------QRLCLHFHQNHQRILLLFHQNH-QRSHQHHQRS---- 673
Query: 171 DGVLPKQQAKKRHPHYDNDEDSLHEDLPKNH--QAKRGRSHY 210
Q ++ H H + + H+ L +H Q + RSH+
Sbjct: 674 ----LLQHRQRSHVH-QSSRNHFHQSLSCHHFLQFQSTRSHF 784
>TC206766 similar to PIR|T47576|T47576 FKBP12 interacting protein (FIP37) -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(74%)
Length = 1952
Score = 32.7 bits (73), Expect = 0.23
Identities = 29/135 (21%), Positives = 49/135 (35%), Gaps = 1/135 (0%)
Frame = +3
Query: 162 HYDDDDDLPDGVLPKQQAKKR-HPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGV 220
H+DDD D DG + KR P YD DED ++ + V ++ +
Sbjct: 201 HFDDDFDFGDGFGGRHSGNKRSSPDYD-DEDYDNDPFAPKKAITKAEEASGVTTGMILSL 377
Query: 221 LPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHP 280
K N+ ++ ++ H + QP + G+ P+ +
Sbjct: 378 RESLQNCKDTLATCQNELEAAKSEIQSWHSTLKNQP------SILAGITPEPKMLINYLQ 539
Query: 281 HYDNDEDSLHEDLPK 295
+ E+SL E L K
Sbjct: 540 ALKSSEESLREQLEK 584
Score = 30.4 bits (67), Expect = 1.1
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Frame = +3
Query: 138 SHYDDDDSLDEVLPKQHHA-KRGHPHYDDDDDLPDGVLPKQQAKK 181
+H+DDD + +H KR P YDD+D D PK+ K
Sbjct: 198 THFDDDFDFGDGFGGRHSGNKRSSPDYDDEDYDNDPFAPKKAITK 332
Score = 27.3 bits (59), Expect = 9.6
Identities = 17/43 (39%), Positives = 22/43 (50%), Gaps = 2/43 (4%)
Frame = +3
Query: 67 HYDDDYSLDEIFPNRRQKHQAHKRHY-HYDDED-SLDSLLPKQ 107
H+DDD+ + F R H +KR YDDED D PK+
Sbjct: 201 HFDDDFDFGDGFGGR---HSGNKRSSPDYDDEDYDNDPFAPKK 320
>TC220068 similar to GB|AAL31246.1|16974485|AY061919 AT4g32480/F8B4_180
{Arabidopsis thaliana;} , partial (10%)
Length = 654
Score = 32.0 bits (71), Expect = 0.39
Identities = 16/83 (19%), Positives = 34/83 (40%)
Frame = +2
Query: 151 PKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHY 210
P +HH +D DD+ + +L + + H + E+ +D + R+ +
Sbjct: 86 PLEHHRDNADVEFDFLDDIGEALLAQSASSDECGHPNEMEEQFDQDEDHRDTVEENRTFW 265
Query: 211 DVEDNLLDGVLPKQPQAKKRHPH 233
D + LL + + ++ R H
Sbjct: 266 DKQHQLLQASICRTSSSESRIRH 334
>TC226273 similar to GB|AAP37745.1|30725446|BT008386 At5g61156 {Arabidopsis
thaliana;} , partial (34%)
Length = 1407
Score = 31.6 bits (70), Expect = 0.51
Identities = 36/185 (19%), Positives = 63/185 (33%), Gaps = 13/185 (7%)
Frame = +2
Query: 20 RARQSAGAFTSVSQARRGHPRYDDDHSL-----DEFLADRRRKKHLALRRHPHYDDDYSL 74
R RQ + F + Y D+H D+ + R +K + + D
Sbjct: 446 RRRQLSPGFLEDALDEEDEADYYDNHRSQRRFEDDLEVEARAEKRIMNAKKSQGPKDIPR 625
Query: 75 DEIFPNRRQKHQA------HKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLP 128
FP + + Y+ E+ D P + + E Y+DE+ +E
Sbjct: 626 KSSFPPAKSSRNPMGYPDDEREESEYETEEEEDERPPSRKRDEDTEPEYEDEEEEEEHYE 805
Query: 129 KQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPH--Y 186
+ Q ++++ + + AKR DDD P++ R Y
Sbjct: 806 EAEQVNDASDDGEEEEPKQKSKEFRGSAKRKGFESDDDSS------PRKITTHRRMAVVY 967
Query: 187 DNDED 191
D+DED
Sbjct: 968 DSDED 982
>TC217533 similar to UP|Q9LUS2 (Q9LUS2) Chloroplast outer envelope
protein-like (Chloroplast outer envelope
membrane-associated protein Toc120), partial (35%)
Length = 1633
Score = 31.6 bits (70), Expect = 0.51
Identities = 31/108 (28%), Positives = 49/108 (44%), Gaps = 6/108 (5%)
Frame = +1
Query: 100 LDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRG 159
L +LL + Q + ++ DEDSLD+ L + ++ H DD + L K K
Sbjct: 148 LSTLLQSRPQLKLPEEQFGDEDSLDDDLDESSESDDENEH--DDLPPFKPLTKAQVEKLS 321
Query: 160 HPH---YDDDDDLPDGVLPKQQ---AKKRHPHYDNDEDSLHEDLPKNH 201
H Y D+ + + +L K+Q KKR +S +DLP +H
Sbjct: 322 KAHKKAYFDELEYREKLLMKKQLKEEKKRRKMLKKMAESA-KDLPSDH 462
>TC225325 similar to UP|R17D_ARATH (Q9LZ17) 40S ribosomal protein S17-4,
partial (86%)
Length = 763
Score = 31.6 bits (70), Expect = 0.51
Identities = 19/92 (20%), Positives = 35/92 (37%), Gaps = 10/92 (10%)
Frame = +3
Query: 124 DEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPD----------GV 173
D LP +++A R H+ ++ ++ H + HP LP G+
Sbjct: 204 DPRLPHKQKASRRSGHHPFQEAAEQDRGFLHASHEAHPEGSRSRHLPQAPGGGARAPHGL 383
Query: 174 LPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKR 205
P++ + PH + H P+N + R
Sbjct: 384 RPRRLRHQHRPHRSRQRNP*HASFPRNQRHPR 479
>CA784384
Length = 409
Score = 31.2 bits (69), Expect = 0.66
Identities = 17/45 (37%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Frame = +3
Query: 107 QHQAERGHTRYDDEDSLD--EVLPKQRQAKRGRSHYDDDDSLDEV 149
+ + R H DDED +D E+L KQ + R DDD DE+
Sbjct: 159 KQKKSRDHVTKDDEDGVDAFELLFKQLEEDLKRGDLSDDDGEDEI 293
>TC225458 UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37, complete
Length = 1797
Score = 31.2 bits (69), Expect = 0.66
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 4/55 (7%)
Frame = +2
Query: 217 LDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPR----YDDDDDLPDG 267
L+ VLP +P ++ D ++ D+ + R Q + YD+DDD+P G
Sbjct: 1175 LEAVLPPKPSSQLTDMELDECEETTLHDVNMEEETRRKQQQAQEAYDEDDDMPGG 1339
Score = 27.7 bits (60), Expect = 7.3
Identities = 30/115 (26%), Positives = 47/115 (40%), Gaps = 8/115 (6%)
Frame = +2
Query: 75 DEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAK 134
DE P ++ K + H+ E DSL P Q +A L+ VLP + ++
Sbjct: 1070 DEGMPMYQRPFMKGKLYIHFTVEFP-DSLNPDQVKA------------LEAVLPPKPSSQ 1210
Query: 135 RGRSHYDDDDSL--------DEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKK 181
D+ + +E KQ A+ YD+DDD+P G Q A++
Sbjct: 1211 LTDMELDECEETTLHDVNMEEETRRKQQQAQEA---YDEDDDMPGGAQRVQCAQQ 1366
>TC214767 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (39%)
Length = 713
Score = 31.2 bits (69), Expect = 0.66
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 4/55 (7%)
Frame = +2
Query: 217 LDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPR----YDDDDDLPDG 267
L+ VLP +P ++ D ++ D+ + R Q + YD+DDD+P G
Sbjct: 296 LEAVLPPKPSSQLTDMELDECEETTLHDVNMEEETRRKQQQAQEAYDEDDDMPGG 460
Score = 28.1 bits (61), Expect = 5.6
Identities = 30/115 (26%), Positives = 47/115 (40%), Gaps = 8/115 (6%)
Frame = +2
Query: 75 DEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAK 134
DE P ++ K + H+ E DSL P Q +A L+ VLP + ++
Sbjct: 191 DEGMPMYQRSFMKGKLYIHFTVEFP-DSLNPDQVKA------------LEAVLPPKPSSQ 331
Query: 135 RGRSHYDDDDSL--------DEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKK 181
D+ + +E KQ A+ YD+DDD+P G Q A++
Sbjct: 332 LTDMELDECEETTLHDVNMEEETRRKQQQAQEA---YDEDDDMPGGAQRVQCAQQ 487
>BM308153 similar to GP|6960465|gb|A ORF4 hoar {Spodoptera exigua
nucleopolyhedrovirus}, partial (2%)
Length = 432
Score = 31.2 bits (69), Expect = 0.66
Identities = 19/66 (28%), Positives = 29/66 (43%), Gaps = 6/66 (9%)
Frame = +3
Query: 62 LRRHPHYDDDYSL--DEI----FPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHT 115
+R H +++ L DEI N + H H H+H DD+D++ H H
Sbjct: 198 VRDHGELEEELELLLDEIPHATSHNLLRHHHHHHHHHHGDDDDAVFVTKQTHHGHAHAHG 377
Query: 116 RYDDED 121
Y D+D
Sbjct: 378 MYYDDD 395
Score = 27.3 bits (59), Expect = 9.6
Identities = 15/42 (35%), Positives = 19/42 (44%), Gaps = 5/42 (11%)
Frame = +3
Query: 153 QHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPH-----YDND 189
+HH H H+ DDD D V +Q H H YD+D
Sbjct: 279 RHHHHHHHHHHGDDD---DAVFVTKQTHHGHAHAHGMYYDDD 395
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,089,365
Number of Sequences: 63676
Number of extensions: 279424
Number of successful extensions: 1330
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 1013
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1279
length of query: 326
length of database: 12,639,632
effective HSP length: 98
effective length of query: 228
effective length of database: 6,399,384
effective search space: 1459059552
effective search space used: 1459059552
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC142222.3


