

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142095.2 + phase: 0
(458 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
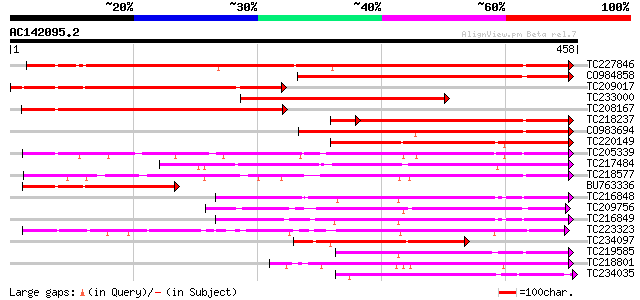
Score E
Sequences producing significant alignments: (bits) Value
TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate S-... 369 e-102
CO984858 300 9e-82
TC209017 similar to UP|Q9M6E7 (Q9M6E7) UDP-glucose:salicylic aci... 286 1e-77
TC233000 similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7 (Fr... 245 3e-65
TC208167 weakly similar to UP|Q9M6E7 (Q9M6E7) UDP-glucose:salicy... 231 4e-61
TC218237 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferas... 215 3e-60
CO983694 206 2e-53
TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferas... 179 3e-45
TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucos... 166 2e-41
TC217484 164 8e-41
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 161 7e-40
BU763336 weakly similar to GP|7385017|gb|A UDP-glucose:salicylic... 156 2e-38
TC216848 similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 8... 144 1e-34
TC209756 141 7e-34
TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransf... 140 9e-34
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 139 3e-33
TC234097 similar to UP|Q9ZR25 (Q9ZR25) UDP-glucose:anthocyanin 5... 139 3e-33
TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltr... 136 2e-32
TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, ... 132 4e-31
TC234035 homologue to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosy... 132 4e-31
>TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate
S-glucosyltransferase, partial (49%)
Length = 1605
Score = 369 bits (947), Expect = e-102
Identities = 200/448 (44%), Positives = 286/448 (63%), Gaps = 6/448 (1%)
Frame = +2
Query: 14 LVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISDGFDK 73
LVLP+PA GH NP+++F+K L + GVK T+ TT +I PN IT+E ISDGFD+
Sbjct: 2 LVLPYPAQGHINPLVQFAKRLASK-GVKATVATTHYTANSI-NAPN--ITVEAISDGFDQ 169
Query: 74 GGVAEAKD-FKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKEFGI 132
G A+ + +L+L F G ++L+ LI C++YDSF PW LDVAK+ GI
Sbjct: 170 AGFAQTNNNVQLFLASFRTNGSRTLSELIRKHQQTPSPSTCIVYDSFFPWVLDVAKQHGI 349
Query: 133 VGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITL--PALPQLQPRDMPSFYFTYEQDPT 190
GA+F T + + +I+ +H G ++ P + + L P LP L R +PSF E P
Sbjct: 350 YGAAFFTNSAAVCNIFCRLHHGFIQLPVKMEHLPLRVPGLPPLDSRALPSFVRFPESYPA 529
Query: 191 FLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKRVKDD 250
++ + ++QFSN++ ADW+ N+F LE EV +++ + +GP +P +LD R+K D
Sbjct: 530 YMAMKLSQFSNLNNADWMFVNTFEALESEVLKGLTELFPA-KMIGPMVPSGYLDGRIKGD 706
Query: 251 EDHSIAQLK--SDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWV 308
+ + + K ++E WL +KP +S VY+SFGSM SL EEQ+EEVA LK+ G FLWV
Sbjct: 707 KGYGASLWKPLTEECSNWLESKPPQSVVYISFGSMVSLTEEQMEEVAWGLKESGVSFLWV 886
Query: 309 VKTSEETKLPKDF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPI 367
++ SE KLP + E + GL+V WC QLE+LAH+A GCFVTHCGWNSTLE+LS+GVP+
Sbjct: 887 LRESEHGKLPLGYRESVKDKGLIVTWCNQLELLAHQATGCFVTHCGWNSTLESLSLGVPV 1066
Query: 368 VAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNV 427
V +P ++DQ DAKFL +IW+VG+ P DEK IVRK + ++M ++ +EI N
Sbjct: 1067VCLPQWADQLPDAKFLDEIWEVGVWPKEDEKGIVRKQEFVQSLKDVME-GQRSQEIRRNA 1243
Query: 428 MQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+WK LA AVG+ GSS K++ +FV+ L
Sbjct: 1244NKWKKLAREAVGEGGSSDKHINQFVDHL 1327
>CO984858
Length = 778
Score = 300 bits (768), Expect = 9e-82
Identities = 139/223 (62%), Positives = 177/223 (79%)
Frame = -2
Query: 233 TVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIE 292
T+GP +P FLDK+ +DD+D+ + Q KS+E +EWL++KPK S VYVSFGSMA+++EEQ+E
Sbjct: 777 TIGPNVPSFFLDKQCEDDQDYGVTQFKSEECVEWLDDKPKGSVVYVSFGSMATMSEEQME 598
Query: 293 EVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLVVAWCPQLEVLAHEAIGCFVTHC 352
EVA CL++C SYFLWVV+ SEE KLPKDFEK +E GLVV WC QL+VLAHEA+GCFVTHC
Sbjct: 597 EVACCLRECSSYFLWVVRASEEIKLPKDFEKITEKGLVVTWCSQLKVLAHEAVGCFVTHC 418
Query: 353 GWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICE 412
GWNS LE L +GVP +AIP +SDQ +AK + D+WK+GIR VDEK IVR++ LK CI E
Sbjct: 417 GWNSILETLCLGVPTIAIPCWSDQRTNAKLIADVWKIGIRTPVDEKNIVRREALKHCIKE 238
Query: 413 IMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
IM ++ KE+ N +QWKTLA RA + GSS++N+IEF N L
Sbjct: 237 IM---DRDKEMKTNAIQWKTLAVRATAEGGSSYENIIEFTNHL 118
>TC209017 similar to UP|Q9M6E7 (Q9M6E7) UDP-glucose:salicylic acid
glucosyltransferase, partial (7%)
Length = 722
Score = 286 bits (732), Expect = 1e-77
Identities = 136/223 (60%), Positives = 176/223 (77%)
Frame = +1
Query: 1 MENKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNN 60
ME K+++ ++ HCLVLP+P GH NPML+FSKLL+ Q GV++TLVTT Y N+ K+P
Sbjct: 64 MEKKSMARRA-HCLVLPYPLQGHINPMLQFSKLLEHQ-GVRITLVTTRFFYNNLQKVPP- 234
Query: 61 SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFM 120
SI +ETISDGFD GG EA K YL++FWQVGP++ A L+ L NDHVDC++YD+F+
Sbjct: 235 SIVLETISDGFDLGGPKEAGGSKAYLDRFWQVGPETFAELLEKLGKSNDHVDCVVYDAFL 414
Query: 121 PWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPS 180
PW LDVAK FGIVGA++LTQN+ +NSIYYHV LGKL+ P +E +I+LPALP+L +DMP+
Sbjct: 415 PWALDVAKRFGIVGAAYLTQNMTVNSIYYHVQLGKLQAPLIEHDISLPALPKLHLKDMPT 594
Query: 181 FYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADW 223
F+F ++DP+ LD VAQFSNI KADWILCN+F EL+KE+ DW
Sbjct: 595 FFF--DEDPSLLDFVVAQFSNIDKADWILCNTFNELDKEIVDW 717
>TC233000 similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7 (Fragment),
partial (58%)
Length = 509
Score = 245 bits (626), Expect = 3e-65
Identities = 108/169 (63%), Positives = 137/169 (80%)
Frame = +1
Query: 187 QDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKR 246
++ T LD V QFSNI KADWILCN+++EL+KE+ DW M+IW FR++GP +P FLDKR
Sbjct: 1 RERTMLDFFVVQFSNIDKADWILCNTYYELDKEIVDWIMEIWPKFRSIGPNIPSLFLDKR 180
Query: 247 VKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFL 306
++D+D+ + + K DE IEWL++KPK S VYVSFGS+A+ +EQ+EE+A CLK+ YFL
Sbjct: 181 YENDQDYGVTEFKRDECIEWLDDKPKGSVVYVSFGSIATFGDEQMEELACCLKESLGYFL 360
Query: 307 WVVKTSEETKLPKDFEKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWN 355
WVV+ SEETKLPK FEKK++ GLVV WC QL+VLAHEAIGCFVTHCGWN
Sbjct: 361 WVVRASEETKLPKGFEKKTKKGLVVTWCSQLKVLAHEAIGCFVTHCGWN 507
>TC208167 weakly similar to UP|Q9M6E7 (Q9M6E7) UDP-glucose:salicylic acid
glucosyltransferase, partial (23%)
Length = 890
Score = 231 bits (590), Expect = 4e-61
Identities = 111/215 (51%), Positives = 149/215 (68%)
Frame = +3
Query: 10 SVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISD 69
+ HCLVLP+PA GH NPML+FSK L Q+ GVKVTLVT +SN+KN+ SI +E+ISD
Sbjct: 60 AAHCLVLPYPAQGHINPMLQFSKRLVQR-GVKVTLVTVVSNWKNMRNKNFTSIEVESISD 236
Query: 70 GFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKE 129
G+D GG+A A+ + Y+ FW+VG Q+ A L+ L + DC+IYD+FMPW LDVAK+
Sbjct: 237 GYDDGGLAAAESLEAYIETFWRVGSQTFAELVQKLAGSSHPPDCVIYDAFMPWVLDVAKK 416
Query: 130 FGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYEQDP 189
FG++GA+F TQ N+IY+HV+ ++ P + E LP LP+L D+PSF Y P
Sbjct: 417 FGLLGATFFTQTCTTNNIYFHVYKKLIELPLTQAEYLLPGLPKLAAGDLPSFLNKYGSYP 596
Query: 190 TFLDIGVAQFSNIHKADWILCNSFFELEKEVADWT 224
+ D+ V QF NI KADW+L NSF+ELE+ V+D T
Sbjct: 597 GYFDVVVNQFVNIDKADWVLANSFYELEQGVSDLT 701
>TC218237 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (57%)
Length = 1176
Score = 215 bits (547), Expect(2) = 3e-60
Identities = 98/175 (56%), Positives = 134/175 (76%)
Frame = +3
Query: 281 GSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLVVAWCPQLEVL 340
G+ A LNEEQ EE+A L D GSYF+WV++ ++ KLPK+F SE GL+V+WCPQL+VL
Sbjct: 126 GA*AGLNEEQTEELAWGLGDSGSYFMWVIRDCDKGKLPKEFADTSEKGLIVSWCPQLQVL 305
Query: 341 AHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQI 400
HEA+GCF+THCGWNSTLEALS+GVP++A+PL++DQ +AK L D+WK+G++ + DEK+I
Sbjct: 306 THEALGCFLTHCGWNSTLEALSLGVPVIAMPLWTDQITNAKLLKDVWKIGVKAVADEKEI 485
Query: 401 VRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
VR++ + CI EI+ +EKG EI N ++WK LA V + G+S KN+ EFV L
Sbjct: 486 VRRETITHCIKEILE-TEKGNEIKKNAIKWKNLAKSYVDEGGNSDKNIAEFVEEL 647
Score = 35.4 bits (80), Expect(2) = 3e-60
Identities = 15/24 (62%), Positives = 19/24 (78%)
Frame = +2
Query: 260 SDESIEWLNNKPKRSAVYVSFGSM 283
S+ I+WL+ KPK S VYVSFGS+
Sbjct: 62 SEACIKWLDEKPKGSVVYVSFGSI 133
>CO983694
Length = 858
Score = 206 bits (523), Expect = 2e-53
Identities = 103/226 (45%), Positives = 146/226 (64%), Gaps = 4/226 (1%)
Frame = -1
Query: 234 VGPCLPYTFLDKRVKDDEDHSIAQLKSDES-IEWLNNKPKRSAVYVSFGSMASLNEEQIE 292
+GP +P LDK V +D D+ + D S I WL KP S +Y+SFGSM + +Q+E
Sbjct: 849 IGPTVPXXHLDKAVPNDTDNXXNXFQVDSSAISWLRQKPAGSVIYISFGSMVCFSSQQME 670
Query: 293 EVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSE---NGLVVAWCPQLEVLAHEAIGCFV 349
E+A L G FLWV+ E LPK+ ++ GL+V W PQLEVL++ A+GCF
Sbjct: 669 EIALGLMATGFNFLWVIPDLERKNLPKELGEEINACGRGLIVNWTPQLEVLSNHAVGCFF 490
Query: 350 THCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDC 409
THCGWNSTLEAL +GVP+VA+P ++DQ +AKF+ D+WKVGIR +E IV ++ +++C
Sbjct: 489 THCGWNSTLEALCLGVPMVALPQWTDQPTNAKFVEDVWKVGIRVKENENGIVTREEVENC 310
Query: 410 ICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
I +M + G+E+ N +WK LA AV + G+S N+ EF+N+L
Sbjct: 309 IRVVME-KDLGREMRINAKKWKELAIEAVSQGGTSDNNINEFINNL 175
>TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferase like
protein, partial (27%)
Length = 820
Score = 179 bits (453), Expect = 3e-45
Identities = 87/201 (43%), Positives = 132/201 (65%), Gaps = 5/201 (2%)
Frame = +2
Query: 260 SDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPK 319
S++ +EWL+++P+ S VYVSFG++A L + Q++E+A L D G FLWV++ + +
Sbjct: 5 SNDYVEWLDSQPELSVVYVSFGTLAVLADRQMKELARALLDSGYLFLWVIRDMQGIE-DN 181
Query: 320 DFEKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGID 379
E+ + G +V WC Q+EVL+H ++GCFVTHCGWNST+E+L GVP+VA P ++DQG +
Sbjct: 182 CREELEQRGKIVKWCSQVEVLSHGSLGCFVTHCGWNSTMESLGSGVPMVAFPQWTDQGTN 361
Query: 380 AKFLVDIWKVGIRPLVDEK-----QIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLA 434
AK + D+WK G+R VD+K IV + ++ C+ +M KG+E N +WK LA
Sbjct: 362 AKMVQDVWKTGVR--VDDKVNVEEGIVEAEEIRKCLDVVMGSGGKGQEFRRNADKWKCLA 535
Query: 435 TRAVGKDGSSHKNMIEFVNSL 455
AV + GSS NM F++ +
Sbjct: 536 REAVTEGGSSDSNMRTFLHDV 598
>TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose
glucosyltransferase, partial (55%)
Length = 1537
Score = 166 bits (421), Expect = 2e-41
Identities = 138/487 (28%), Positives = 232/487 (47%), Gaps = 42/487 (8%)
Frame = +2
Query: 11 VHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIP----KLPNNSITIET 66
+H ++ PFP GH PM + ++ + GV+ T+VTT N I K I I T
Sbjct: 29 LHIMLFPFPGQGHLIPMSDMARAFNGR-GVRTTIVTTPLNVATIRGTIGKETETDIEILT 205
Query: 67 ISDGFDKGGVAE---------AKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYD 117
+ + G+ E + D L K ++ L HL+ CLI
Sbjct: 206 VKFPSAEAGLPEGCENTESIPSPDLVLTFLKAIRMLEAPLEHLL-----LQHRPHCLIAS 370
Query: 118 SFMPWCLDVAKEFGI-------VGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPAL 170
+F PW A + I G L + + H ++ PF+ +P L
Sbjct: 371 AFFPWASHSATKLKIPRLVFHGTGVFALCASECVRLYQPHKNVSSDTDPFI-----IPHL 535
Query: 171 P---QLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADW-ILCNSFFELEKEVADWTMK 226
P Q+ +P + T T L + + A + ++ NSF+ELE+ AD+ K
Sbjct: 536 PGDIQMTRLLLPDYAKTDGDGETGLTRVLQEIKESELASYGMIVNSFYELEQVYADYYDK 715
Query: 227 IWSNFRT-----VGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFG 281
+ +GP L DK + + A + + ++WL++K S VYV FG
Sbjct: 716 QLLQVQGRRAWYIGP-LSLCNQDKGKRGKQ----ASVDQGDILKWLDSKKANSVVYVCFG 880
Query: 282 SMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETK---LPKDFEKKSEN---GLVV-AWC 334
S+A+ +E Q+ E+A L+D G F+WVV+ S++ LP+ FE ++ + G+++ W
Sbjct: 881 SIANFSETQLREIARGLEDSGQQFIWVVRRSDKDDKGWLPEGFETRTTSEGRGVIIWGWA 1060
Query: 335 PQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPL 394
PQ+ +L H+A+G FVTHCGWNSTLEA+S GVP++ P+ ++Q + KF+ DI ++G+ P+
Sbjct: 1061PQVLILDHQAVGAFVTHCGWNSTLEAVSAGVPMLTWPVSAEQFYNEKFVTDILQIGV-PV 1237
Query: 395 VDEK------QIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNM 448
+K + + L+ + IM + E+ + + N + +AT A+ +GSS+ +
Sbjct: 1238GVKKWNRIVGDNITSNALQKALHRIM-IGEEAEPMRNRAHKLAQMATTALQHNGSSYCHF 1414
Query: 449 IEFVNSL 455
+ L
Sbjct: 1415THLIQHL 1435
>TC217484
Length = 1651
Score = 164 bits (415), Expect = 8e-41
Identities = 104/346 (30%), Positives = 179/346 (51%), Gaps = 12/346 (3%)
Frame = +2
Query: 122 WCLDVAKEFGIVGASFLTQNLVMNSIYYHV-----HLGKL--KPPFVEQEITLPALPQLQ 174
W + VA I A+F T + S+ +H+ H G K Q +P +
Sbjct: 233 WPIAVANRRNIPVAAFWTMSASFYSMLHHLDVFARHRGLTVDKDTMDGQAENIPGISSAH 412
Query: 175 PRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWS-NFRT 233
D+ + +E D + + + S + +A+++L + ELE E + I+
Sbjct: 413 LADLRTVL--HENDQRVMQLALECISKVPRANYLLLTTVQELEAETIESLKAIFPFPVYP 586
Query: 234 VGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEE 293
+GP +PY L + ++ DHS + I+WL+++P S +Y+SFGS S++ Q+++
Sbjct: 587 IGPAIPYLELGQNPLNN-DHS------HDYIKWLDSQPPESVLYISFGSFLSVSTTQMDQ 745
Query: 294 VAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLVVAWCPQLEVLAHEAIGCFVTHCG 353
+ L +LWV + + EK + G+VV WC QL+VL+H ++G F +HCG
Sbjct: 746 IVEALNSSEVRYLWVARANASFLK----EKCGDKGMVVPWCDQLKVLSHSSVGGFWSHCG 913
Query: 354 WNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR---PLVDEKQIVRKDPLKDCI 410
WNSTLEAL GVP++ PL+ DQ ++ +VD WK G + +D + IV K+ +++ +
Sbjct: 914 WNSTLEALFAGVPMLTFPLFLDQVPNSSQIVDEWKNGSKVETSKLDSEVIVAKEKIEELV 1093
Query: 411 CEIMSM-SEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
M + S++GKEI + + K + RA+ GSS+ N+ F+ +
Sbjct: 1094KRFMDLQSQEGKEIRDRAREIKVMCLRAIAAGGSSYGNLDAFIRDI 1231
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 161 bits (407), Expect = 7e-40
Identities = 137/486 (28%), Positives = 219/486 (44%), Gaps = 42/486 (8%)
Frame = +1
Query: 12 HCLVLPFPAHGHTNPMLEFSK-LLQQQEGVKVTLV------TTISNYKNIPKLPNN---- 60
H V+P P H P+LEFSK LL +T + S+ + LP
Sbjct: 4 HVAVVPSPGFTHLVPILEFSKRLLHLHPEFHITCFIPSVGSSPTSSKAYVQTLPPTITSI 183
Query: 61 ---SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYD 117
IT++ +SD A +L +N + L L + V L+ D
Sbjct: 184 FLPPITLDHVSDPS-----VLALQIELSVNLSLPYIREELKSLCSRAK-----VVALVVD 333
Query: 118 SFMPWCLDVAKEFGIVGASFLTQNLVMNSIY-YHVHLGKL---KPPFVEQEITLPALPQL 173
F L+ AKE ++ +L Q+ ++ S+Y Y L ++ + +++ I +P +
Sbjct: 334 VFANGALNFAKELNLLSYIYLPQSAMLLSLYFYSTKLDEILSSESRELQKPIDIPGCVPI 513
Query: 174 QPRDMPSFYFTYEQDPTFLDIGVAQF----SNIHKADWILCNSFFELEK---EVADWTMK 226
+D+P + +G F H D + N+F ELE + +K
Sbjct: 514 HNKDLPLPFHDLS------GLGYKGFLERSKRFHVPDGVFMNTFLELESGAIRALEEHVK 675
Query: 227 IWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASL 286
VGP + + SI E + WL+ + S +YVSFGS +L
Sbjct: 676 GKPKLYPVGPII------------QMESIGHENGVECLTWLDKQEPNSVLYVSFGSGGTL 819
Query: 287 NEEQIEEVAHCLKDCGSYFLWVVKTSE----------ETKLPKDF------EKKSENGLV 330
++EQ E+A L+ G FLWVV+ ETK P +F E+ + GLV
Sbjct: 820 SQEQFNELAFGLELSGKKFLWVVRAPSGVVSAGYLCAETKDPLEFLPHGFLERTKKQGLV 999
Query: 331 V-AWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKV 389
V +W PQ++VL H A G F++HCGWNS LE++ GVP++ PL+++Q ++A + D KV
Sbjct: 1000VPSWAPQIQVLGHSATGGFLSHCGWNSVLESVVQGVPVITWPLFAEQSLNAAMIADDLKV 1179
Query: 390 GIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMI 449
+RP V+E +V ++ + + +M ++ EI + K A A+ +DGSS K +
Sbjct: 1180ALRPKVNESGLVEREEIAKVVRGLMG-DKESLEIRKRMGLLKIAAANAIKEDGSSTKTLS 1356
Query: 450 EFVNSL 455
E SL
Sbjct: 1357EMATSL 1374
>BU763336 weakly similar to GP|7385017|gb|A UDP-glucose:salicylic acid
glucosyltransferase {Nicotiana tabacum}, partial (6%)
Length = 427
Score = 156 bits (394), Expect = 2e-38
Identities = 77/127 (60%), Positives = 94/127 (73%)
Frame = +2
Query: 11 VHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISDG 70
VHC+VL +PA GH NPM F KLLQQQ GVKVTLVTT+S KN+ +P SI +ETISDG
Sbjct: 53 VHCVVLAYPAQGHINPMHNFCKLLQQQ-GVKVTLVTTLSYSKNLQNIPA-SIALETISDG 226
Query: 71 FDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKEF 130
FD G AE+ ++K YL +FWQVGP++LA L+ L D VDC++Y+SF PW L+VAK F
Sbjct: 227 FDNRGFAESGNWKAYLERFWQVGPKTLAELLEKLGRSGDPVDCVVYNSFFPWALEVAKRF 406
Query: 131 GIVGASF 137
GIVG F
Sbjct: 407 GIVGGVF 427
>TC216848 similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8, partial
(39%)
Length = 1113
Score = 144 bits (362), Expect = 1e-34
Identities = 83/296 (28%), Positives = 158/296 (53%), Gaps = 7/296 (2%)
Frame = +1
Query: 167 LPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMK 226
+P L + +D+P F T + + L + + A + N+F ELE++ +
Sbjct: 10 IPGLQNYRLKDLPDFLRTTDPNDFMLHFFIEVAEKVPSASAVAFNTFHELERDAINALPS 189
Query: 227 IWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDES--IEWLNNKPKRSAVYVSFGSMA 284
++ + ++GP P +FLD+ + L +++ ++WL +K RS VYV+FGS+
Sbjct: 190 MFPSLYSIGP-FP-SFLDQSPHKQVPSLGSNLWKEDTGCLDWLESKEPRSVVYVNFGSIT 363
Query: 285 SLNEEQIEEVAHCLKDCGSYFLWVVKTS----EETKLPKDFEKKS-ENGLVVAWCPQLEV 339
++ EQ+ E A L + FLW+++ L +F ++ + L+ +WCPQ +V
Sbjct: 364 VMSAEQLLEFAWGLANSKKPFLWIIRPDLVIGGSVILSSEFVNETRDRSLIASWCPQEQV 543
Query: 340 LAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQ 399
L H +IG F+THCGWNST E++ GVP++ P ++DQ + +++ + W++G+ +D
Sbjct: 544 LNHPSIGVFLTHCGWNSTTESICAGVPMLCWPFFADQPTNCRYICNEWEIGME--IDTN- 714
Query: 400 IVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+++ L+ + E+M + EKGK++ M+ K A G S+ N+ + + +
Sbjct: 715 -AKREELEKLVNELM-VGEKGKKMGQKTMELKKKAEEETRPGGGSYMNLDKLIKEV 876
>TC209756
Length = 1356
Score = 141 bits (355), Expect = 7e-34
Identities = 86/301 (28%), Positives = 148/301 (48%), Gaps = 6/301 (1%)
Frame = +1
Query: 159 PFVEQEITL-PALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELE 217
P QEI L P +P + + P F + F D V + + +W LCN+ ++LE
Sbjct: 40 PTKTQEIQLSPNMPLIDTENFPWRGF----NKIFFDHLVQEMKTLELGEWWLCNTTYDLE 207
Query: 218 KEVADWTMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVY 277
+ K F +GP + + D S + +EWL+ +P +S +Y
Sbjct: 208 PGAFSVSPK----FLPIGPLM---------ESDNSKSAFWEEDTTCLEWLDQQPPQSVIY 348
Query: 278 VSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETK-----LPKDFEKKSENGLVVA 332
VSFGS+A ++ Q +E+A L F+WVV+ + K DF G +V
Sbjct: 349 VSFGSLAVMDPNQFKELALALDLLDKPFIWVVRPCNDNKENVNAYAHDFH--GSKGKIVG 522
Query: 333 WCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR 392
W PQ ++L H A+ F++HCGWNSTLE + GVP + P +DQ +D ++ D+WK+G+
Sbjct: 523 WAPQKKILNHPALASFISHCGWNSTLEGICAGVPFLCWPCATDQYLDKSYICDVWKIGLG 702
Query: 393 PLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFV 452
DE I+ ++ ++ + +++ ++I ++ K + + + G S KN+ F+
Sbjct: 703 LDKDENGIISREEIRKKVDQLL----VDEDIKARSLKLKDMTINNILEGGQSSKNLNFFM 870
Query: 453 N 453
+
Sbjct: 871 D 873
>TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8,
partial (47%)
Length = 1125
Score = 140 bits (354), Expect = 9e-34
Identities = 85/300 (28%), Positives = 157/300 (52%), Gaps = 11/300 (3%)
Frame = +2
Query: 167 LPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMK 226
+P + + +D+P F T + + L + + I + IL N+F LE +V +
Sbjct: 47 IPGMKNFRLKDIPDFIRTTDLNNVMLQFFIEVANKIQRNTTILFNTFDGLESDVMN---A 217
Query: 227 IWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSD------ESIEWLNNKPKRSAVYVSF 280
+ S F ++ P P+ L + + L S+ E +EWL +K RS VYV+F
Sbjct: 218 LSSMFPSLYPIGPFPLL---LNQSPQSHLTSLGSNLWNEDLECLEWLESKESRSVVYVNF 388
Query: 281 GSMASLNEEQIEEVAHCLKDCGSYFLWVVKTS----EETKLPKDFEKKS-ENGLVVAWCP 335
GS+ ++ EQ+ E A L + FLW+++ L +F ++ + L+ +WCP
Sbjct: 389 GSITVMSAEQLLEFAWGLANSKKPFLWIIRPDLVIGGSVILSSEFVSETRDRSLIASWCP 568
Query: 336 QLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLV 395
Q +VL H +IG F+THCGWNST E++ GVP++ P +++Q + +++ + W++G+ +
Sbjct: 569 QEQVLNHPSIGVFLTHCGWNSTTESVCAGVPMLCWPFFAEQPTNCRYICNEWEIGME--I 742
Query: 396 DEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
D +++ ++ + E+M + EKGK++ VM+ K A G S+ N+ + + +
Sbjct: 743 DTS--AKREEVEKLVNELM-VGEKGKKMREKVMELKRKAEEVTKPGGCSYMNLDKVIKEV 913
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 139 bits (350), Expect = 3e-33
Identities = 117/475 (24%), Positives = 226/475 (46%), Gaps = 33/475 (6%)
Frame = +1
Query: 11 VHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISDG 70
V +++PFPA GH N +L S+L+ + V V T+++ + + +NSI+
Sbjct: 40 VMAVLIPFPAQGHLNQLLHLSRLILSHN-IPVHYVGTVTHIRQVTLRDHNSIS-NIHFHA 213
Query: 71 FDKGGVA--------EAKDFKLYLNKFWQVGP---QSLAHLINNLNARNDHVDCLIYDSF 119
F+ E DF +L ++ + + L+++L+++ V +I+DS
Sbjct: 214 FEVPSFVSPPPNPNNEETDFPAHLLPSFEASSHLREPVRKLLHSLSSQAKRV-IVIHDSV 390
Query: 120 MPWCLDVAKEF-GIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDM 178
M A + +F + +++Y +G+ P V+ + +P +P ++
Sbjct: 391 MASVAQDATNMPNVENYTFHSTCTFGTAVFYWDKMGR---PLVDGML-VPEIPSMEG--- 549
Query: 179 PSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWT------MKIWSNFR 232
FT + F++ +AQ D + N+ +E +W K+W+
Sbjct: 550 ---CFTTD----FMNFMIAQRDFRKVNDGNIYNTSRAIEGAYIEWMERFTGGKKLWA--- 699
Query: 233 TVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIE 292
+GP P F KD ++ +EWL+ + S +YVSFG+ + EEQI+
Sbjct: 700 -LGPFNPLAF---EKKDSKERHFC-------LEWLDKQDPNSVLYVSFGTTTTFKEEQIK 846
Query: 293 EVAHCLKDCGSYFLWVVKTSEE-----------TKLPKDFEKKSEN-GLVVA-WCPQLEV 339
++A L+ F+WV++ +++ + +FE++ E GLVV W PQLE+
Sbjct: 847 KIATGLEQSKQKFIWVLRDADKGDIFDGSEAKWNEFSNEFEERVEGMGLVVRDWAPQLEI 1026
Query: 340 LAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVG--IRPLVDE 397
L+H + G F++HCGWNS LE++S+GVPI A P++SDQ ++ + ++ K+G ++
Sbjct: 1027LSHTSTGGFMSHCGWNSCLESISMGVPIAAWPMHSDQPRNSVLITEVLKIGLVVKNWAQR 1206
Query: 398 KQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFV 452
+V +++ + +M E G ++ ++ K + R++ + G S + F+
Sbjct: 1207NALVSASNVENAVRRLMETKE-GDDMRERAVRLKNVIHRSMDEGGVSRMEIDSFI 1368
>TC234097 similar to UP|Q9ZR25 (Q9ZR25) UDP-glucose:anthocyanin
5-O-glucosyltransferase, partial (30%)
Length = 435
Score = 139 bits (349), Expect = 3e-33
Identities = 71/145 (48%), Positives = 96/145 (65%), Gaps = 3/145 (2%)
Frame = +1
Query: 230 NFRTVGPCLPYTFLDKRVKDDEDHSIAQ---LKSDESIEWLNNKPKRSAVYVSFGSMASL 286
N +GP +P FLD KD D S S++ EWL++KP+ S VYVSFGS L
Sbjct: 7 NMIPIGPLIPSAFLDG--KDPTDTSFGGDIFRPSNDCGEWLDSKPEMSVVYVSFGSFCVL 180
Query: 287 NEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLVVAWCPQLEVLAHEAIG 346
+++Q+EE+A L DCGS FLWV + EE +L E+ + G +V WC Q+EVL+H ++G
Sbjct: 181 SKKQMEELALALLDCGSPFLWVSREKEEEELSCR-EELEQKGKIVNWCSQVEVLSHRSVG 357
Query: 347 CFVTHCGWNSTLEALSIGVPIVAIP 371
CFVTHCGWNST+E+L+ GVP+ A P
Sbjct: 358 CFVTHCGWNSTMESLASGVPMFAFP 432
>TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltransferase ,
partial (50%)
Length = 935
Score = 136 bits (343), Expect = 2e-32
Identities = 65/197 (32%), Positives = 117/197 (58%), Gaps = 5/197 (2%)
Frame = +2
Query: 264 IEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTS----EETKLPK 319
I+WL+ S +YV++GS+ ++E+ ++E A L + FLW+ + E T+LP+
Sbjct: 89 IQWLDQWEPSSVIYVNYGSITVMSEDHLKEFAWGLANSNLPFLWIKRPDLVMGESTQLPQ 268
Query: 320 DF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGI 378
DF ++ + G + WCPQ +VL+H ++G F+THCGWNSTLE +S GVP++ P +++Q
Sbjct: 269 DFLDEVKDRGYITGWCPQEQVLSHPSVGVFLTHCGWNSTLEGISGGVPMIGWPFFAEQQT 448
Query: 379 DAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAV 438
+ +++ W +G +D K V+++ + + E+++ E+GKE+ ++WK A A
Sbjct: 449 NCRYICTTWGIG----MDIKDDVKREEVTTLVKEMIT-GERGKEMRQKCLEWKKKAIEAT 613
Query: 439 GKDGSSHKNMIEFVNSL 455
GSS+ + V +
Sbjct: 614 DMGGSSYNDFHRLVKEV 664
>TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, partial
(60%)
Length = 1281
Score = 132 bits (331), Expect = 4e-31
Identities = 89/275 (32%), Positives = 143/275 (51%), Gaps = 30/275 (10%)
Frame = +1
Query: 211 NSFFELEKEVAD-----WTMKIWSNFRTVGPCLPYTFLDKRVKDD--EDHSIAQLKSDES 263
NSF +LE A+ W K W +GP L R +D E + + ++
Sbjct: 100 NSFHDLEPAYAEQVKNKWGKKAW----IIGP----VSLCNRTAEDKTERGKLPTIDEEKC 255
Query: 264 IEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKT-----SEETK-- 316
+ WLN+K S +YVSFGS+ L EQ++E+A L+ F+WVV+ SE +
Sbjct: 256 LNWLNSKKPNSVLYVSFGSLLRLPSEQLKEIACGLEASEQSFIWVVRNIHNNPSENKENG 435
Query: 317 ----LPKDFE---KKSENGLVV-AWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIV 368
LP+ FE K+ + GLV+ W PQL +L H AI F+THCGWNSTLE++ GVP++
Sbjct: 436 NGNFLPEGFEQRMKEKDKGLVLRGWAPQLLILEHVAIKGFMTHCGWNSTLESVCAGVPMI 615
Query: 369 AIPLYSDQGIDAKFLVDIWKVGIRPLVDE--------KQIVRKDPLKDCICEIMSMSEKG 420
PL ++Q + K + ++ K+G++ E K +V ++ ++ + ++M SE+
Sbjct: 616 TWPLSAEQFSNEKLITEVLKIGVQVGSREWLSWNSEWKDLVGREKVESAVRKLMVESEEA 795
Query: 421 KEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+E+ V A RAV + G+S+ + + L
Sbjct: 796 EEMTTRVKDIAEKAKRAVEEGGTSYADAEALIEEL 900
>TC234035 homologue to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl
transferase, partial (51%)
Length = 825
Score = 132 bits (331), Expect = 4e-31
Identities = 72/201 (35%), Positives = 116/201 (56%), Gaps = 6/201 (2%)
Frame = +2
Query: 264 IEWLNNKPKR-----SAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLP 318
+ WL++K K+ S YVSFG++ + +I VA L+ G FLW +K + LP
Sbjct: 134 LSWLDHKQKQNNGVGSVAYVSFGTVVTPPPHEIVAVAEALEASGVPFLWSLKEHLKGVLP 313
Query: 319 KDF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQG 377
+ F E+ SE+G VVAW PQ +VL H ++G FVTHCG NS E++S GVP++ P + D G
Sbjct: 314 RGFLERTSESGKVVAWAPQTQVLGHGSVGVFVTHCGCNSVFESMSNGVPMICRPFFGDHG 493
Query: 378 IDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRA 437
+ + + D+W++G+R E + KD L C+ ++ + E+GK++ N ++ K A
Sbjct: 494 LTGRMVEDVWEIGVRV---EGGVFTKDGLVKCL-RLVLVEEEGKKMKENAIKVKKTVVDA 661
Query: 438 VGKDGSSHKNMIEFVNSLFQV 458
G G + ++ N+L +V
Sbjct: 662 AGPQGKAAQDF----NTLLEV 712
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,860,677
Number of Sequences: 63676
Number of extensions: 336565
Number of successful extensions: 1996
Number of sequences better than 10.0: 196
Number of HSP's better than 10.0 without gapping: 1870
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1880
length of query: 458
length of database: 12,639,632
effective HSP length: 100
effective length of query: 358
effective length of database: 6,272,032
effective search space: 2245387456
effective search space used: 2245387456
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC142095.2


