

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141923.7 - phase: 0
(406 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
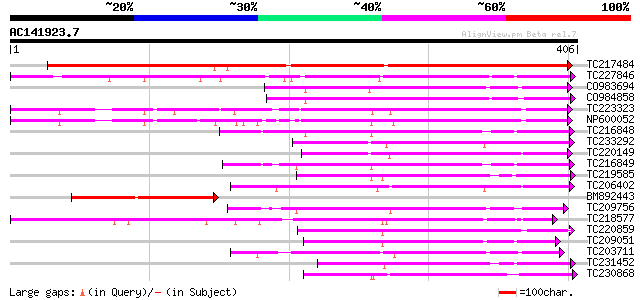
Score E
Sequences producing significant alignments: (bits) Value
TC217484 355 2e-98
TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate S-... 167 1e-41
CO983694 155 4e-38
CO984858 144 5e-35
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 142 2e-34
NP600052 zeatin O-glucosyltransferase [Glycine max] 142 4e-34
TC216848 similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 8... 138 5e-33
TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, pa... 138 5e-33
TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferas... 136 2e-32
TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransf... 135 4e-32
TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltr... 132 3e-31
TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, p... 130 1e-30
BM892443 127 7e-30
TC209756 125 3e-29
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 124 8e-29
TC220859 weakly similar to UP|Q94IF2 (Q94IF2) Glucosyltransferas... 124 1e-28
TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferas... 122 4e-28
TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, pa... 121 6e-28
TC231452 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransf... 120 8e-28
TC230868 weakly similar to UP|Q94AB5 (Q94AB5) AT3g46660/F12A12_1... 120 1e-27
>TC217484
Length = 1651
Score = 355 bits (911), Expect = 2e-98
Identities = 187/415 (45%), Positives = 262/415 (63%), Gaps = 39/415 (9%)
Frame = +2
Query: 28 VSFVVTEEWLSFISSEPKPDNISFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELL 87
++FVVTEEWL FI +EPKPD + + NV+P E + + PAF E V+T+M+APFE LL
Sbjct: 2 ITFVVTEEWLGFIGAEPKPDAVRLAAIPNVVPPERLKAANFPAFYEAVVTEMQAPFERLL 181
Query: 88 DLLDHPPSIIVYDTLLYWAVVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPV-- 145
D L PP+ I+ L W + VANRRNIP A FW M AS +S+ H +F ++ V
Sbjct: 182 DRLQPPPTAILGCVELRWPIAVANRRNIPVAAFWTMSASFYSMLHHLDVFARHRGLTVDK 361
Query: 146 --------KYPGFEWIH---------------------------KAQYLLFSSIYELESQ 170
PG H +A YLL +++ ELE++
Sbjct: 362 DTMDGQAENIPGISSAHLADLRTVLHENDQRVMQLALECISKVPRANYLLLTTVQELEAE 541
Query: 171 AIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHSY-YIEWLDSQPIGSVLYIAQGS 229
I+ LK+ P P+Y IGP IP L +N P N +HS+ YI+WLDSQP SVLYI+ GS
Sbjct: 542 TIESLKAIFPFPVYPIGPAIPYLELGQN---PLNNDHSHDYIKWLDSQPPESVLYISFGS 712
Query: 230 FFSVSSAQIDEIAAALCASNVRFLWIARSEASRLKEICGAHHMGLIMEWCDQLRVLSHPS 289
F SVS+ Q+D+I AL +S VR+LW+AR+ AS LKE CG G+++ WCDQL+VLSH S
Sbjct: 713 FLSVSTTQMDQIVEALNSSEVRYLWVARANASFLKEKCG--DKGMVVPWCDQLKVLSHSS 886
Query: 290 IGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVK-EDVKRDTLV 348
+GGFWSHCGWNST E+L AGVP LT P+++DQ NS +V++WK G +V+ + + +V
Sbjct: 887 VGGFWSHCGWNSTLEALFAGVPMLTFPLFLDQVPNSSQIVDEWKNGSKVETSKLDSEVIV 1066
Query: 349 KKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
K+KI +LV FMDL + ++IR+R+++++ +CL +IA GGS++ + +AFI D+
Sbjct: 1067AKEKIEELVKRFMDLQSQEGKEIRDRAREIKVMCLRAIAAGGSSYGNLDAFIRDI 1231
>TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate
S-glucosyltransferase, partial (49%)
Length = 1605
Score = 167 bits (422), Expect = 1e-41
Identities = 131/459 (28%), Positives = 214/459 (46%), Gaps = 54/459 (11%)
Frame = +2
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+P+P +G+INP++ K L S V+ + ++ NI+ + S+
Sbjct: 8 LPYPAQGHINPLVQFAKRLASKGVKATVATT------HYTANSINAPNITVEAISDGFDQ 169
Query: 61 ELICGRDHPA--FMEDVMTKMEAPFEELLDLLDHPPS---IIVYDTLLYWAVVVANRRNI 115
++ F+ T EL+ PS IVYD+ W + VA + I
Sbjct: 170 AGFAQTNNNVQLFLASFRTNGSRTLSELIRKHQQTPSPSTCIVYDSFFPWVLDVAKQHGI 349
Query: 116 PAALFWPMPASIFSVFLHQH-----IFEQNGHYPVKYPG--------------------- 149
A F+ A++ ++F H + + H P++ PG
Sbjct: 350 YGAAFFTNSAAVCNIFCRLHHGFIQLPVKMEHLPLRVPGLPPLDSRALPSFVRFPESYPA 529
Query: 150 --------FEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSL---IKN 198
F ++ A ++ ++ LES+ + L P + IGP +P L IK
Sbjct: 530 YMAMKLSQFSNLNNADWMFVNTFEALESEVLKGLTELFPAKM--IGPMVPSGYLDGRIKG 703
Query: 199 DP-------KPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVR 251
D KP S WL+S+P SV+YI+ GS S++ Q++E+A L S V
Sbjct: 704 DKGYGASLWKPLTEECS---NWLESKPPQSVVYISFGSMVSLTEEQMEEVAWGLKESGVS 874
Query: 252 FLWIAR-SEASRL----KEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESL 306
FLW+ R SE +L +E GLI+ WC+QL +L+H + G F +HCGWNST ESL
Sbjct: 875 FLWVLRESEHGKLPLGYRE--SVKDKGLIVTWCNQLELLAHQATGCFVTHCGWNSTLESL 1048
Query: 307 VAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGE 366
GVP + LP + DQ ++K + E W+VG KED K +V+K + V+ + + M +G+
Sbjct: 1049SLGVPVVCLPQWADQLPDAKFLDEIWEVGVWPKEDEK--GIVRKQEFVQSLKDVM--EGQ 1216
Query: 367 LTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVMH 405
+++IR + K +K+ ++ GGS+ N F+ +M+
Sbjct: 1217RSQEIRRNANKWKKLAREAVGEGGSSDKHINQFVDHLMN 1333
>CO983694
Length = 858
Score = 155 bits (391), Expect = 4e-38
Identities = 85/232 (36%), Positives = 136/232 (57%), Gaps = 11/232 (4%)
Frame = -1
Query: 183 IYTIGPTIPKFSLIKNDPKPSNTNHSYY------IEWLDSQPIGSVLYIAQGSFFSVSSA 236
I IGPT+P L K P ++ + + I WL +P GSV+YI+ GS SS
Sbjct: 858 ILMIGPTVPXXHLDKAVPNDTDNXXNXFQVDSSAISWLRQKPAGSVIYISFGSMVCFSSQ 679
Query: 237 QIDEIAAALCASNVRFLWIA-----RSEASRLKEICGAHHMGLIMEWCDQLRVLSHPSIG 291
Q++EIA L A+ FLW+ ++ L E A GLI+ W QL VLS+ ++G
Sbjct: 678 QMEEIALGLMATGFNFLWVIPDLERKNLPKELGEEINACGRGLIVNWTPQLEVLSNHAVG 499
Query: 292 GFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKD 351
F++HCGWNST E+L GVP + LP + DQP N+K + + WKVG RVKE+ + +V ++
Sbjct: 498 CFFTHCGWNSTLEALCLGVPMVALPQWTDQPTNAKFVEDVWKVGIRVKEN--ENGIVTRE 325
Query: 352 KIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
++ + M+ D L R++R +KK +++ + +++ GG++ + N FI+++
Sbjct: 324 EVENCIRVVMEKD--LGREMRINAKKWKELAIEAVSQGGTSDNNINEFINNL 175
>CO984858
Length = 778
Score = 144 bits (364), Expect = 5e-35
Identities = 76/228 (33%), Positives = 132/228 (57%), Gaps = 7/228 (3%)
Frame = -2
Query: 185 TIGPTIPKFSLIKNDPKPSNTNHSYY-----IEWLDSQPIGSVLYIAQGSFFSVSSAQID 239
TIGP +P F L K + + + +EWLD +P GSV+Y++ GS ++S Q++
Sbjct: 777 TIGPNVPSFFLDKQCEDDQDYGVTQFKSEECVEWLDDKPKGSVVYVSFGSMATMSEEQME 598
Query: 240 EIAAALCASNVRFLWIAR-SEASRL-KEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHC 297
E+A L + FLW+ R SE +L K+ GL++ WC QL+VL+H ++G F +HC
Sbjct: 597 EVACCLRECSSYFLWVVRASEEIKLPKDFEKITEKGLVVTWCSQLKVLAHEAVGCFVTHC 418
Query: 298 GWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLV 357
GWNS E+L GVP + +P + DQ N+K++ + WK+G R D K +V+++ + +
Sbjct: 417 GWNSILETLCLGVPTIAIPCWSDQRTNAKLIADVWKIGIRTPVDEK--NIVRREALKHCI 244
Query: 358 HEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVMH 405
E MD D ++++ + + + + + + A GGS++ + F + ++H
Sbjct: 243 KEIMDRD----KEMKTNAIQWKTLAVRATAEGGSSYENIIEFTNHLLH 112
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 142 bits (359), Expect = 2e-34
Identities = 119/464 (25%), Positives = 209/464 (44%), Gaps = 61/464 (13%)
Frame = +1
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTE--------------------EWLSFI 40
+PFP +G++N +++L +L++S+N +H VT E SF+
Sbjct: 55 IPFPAQGHLNQLLHLSRLILSHNIPVHYVGTVTHIRQVTLRDHNSISNIHFHAFEVPSFV 234
Query: 41 SSEPKPDNISFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPS--IIV 98
S P P+N ++++PS + + + P +LL L I++
Sbjct: 235 SPPPNPNNEETDFPAHLLPSF------------EASSHLREPVRKLLHSLSSQAKRVIVI 378
Query: 99 YDTLLYWAVVVANRRNIP---------------AALFWP-MPASIFSVFLHQHIFEQNGH 142
+D+++ A V + N+P A +W M + L I G
Sbjct: 379 HDSVM--ASVAQDATNMPNVENYTFHSTCTFGTAVFYWDKMGRPLVDGMLVPEIPSMEGC 552
Query: 143 YPVKYPGFEWIHKAQYL------LFSSIYELESQAIDVLKSKLP-LPIYTIGPTIPKFSL 195
+ + F I + + ++++ +E I+ ++ ++ +GP F+
Sbjct: 553 FTTDFMNF-MIAQRDFRKVNDGNIYNTSRAIEGAYIEWMERFTGGKKLWALGP----FNP 717
Query: 196 IKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWI 255
+ + K S H + +EWLD Q SVLY++ G+ + QI +IA L S +F+W+
Sbjct: 718 LAFEKKDSKERH-FCLEWLDKQDPNSVLYVSFGTTTTFKEEQIKKIATGLEQSKQKFIWV 894
Query: 256 ARS---------EASRLKEICGAHH-----MGLIM-EWCDQLRVLSHPSIGGFWSHCGWN 300
R ++ E MGL++ +W QL +LSH S GGF SHCGWN
Sbjct: 895 LRDADKGDIFDGSEAKWNEFSNEFEERVEGMGLVVRDWAPQLEILSHTSTGGFMSHCGWN 1074
Query: 301 STKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEF 360
S ES+ GVP P++ DQP NS ++ E K+G VK +R+ LV + V
Sbjct: 1075SCLESISMGVPIAAWPMHSDQPRNSVLITEVLKIGLVVKNWAQRNALVSASNVENAVRRL 1254
Query: 361 MDL-DGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
M+ +G+ D+RER+ +L+ + S+ GG + + ++FI+ +
Sbjct: 1255METKEGD---DMRERAVRLKNVIHRSMDEGGVSRMEIDSFIAHI 1377
>NP600052 zeatin O-glucosyltransferase [Glycine max]
Length = 1395
Score = 142 bits (357), Expect = 4e-34
Identities = 132/472 (27%), Positives = 210/472 (43%), Gaps = 69/472 (14%)
Frame = +1
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTE-----------------------EWL 37
+PFP +G++N +++L + + S+N +H T E
Sbjct: 49 IPFPAQGHLNQLLHLARHIFSHNIPVHYVGTATHIRQATLRDHNSNISNIIIHFHAFEVP 228
Query: 38 SFISSEPKPDNISFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPS-- 95
F+S P P+N S+++PS + + P LL L
Sbjct: 229 PFVSPPPNPNNEETDFPSHLLPSF------------KASSHLREPVRNLLQSLSSQAKRV 372
Query: 96 IIVYDTLLYWAVVVANRRNIPAALFWPMPASI-FSVFLHQHIFEQNGHYPVK-------Y 147
I+++D+L+ A V + N+P + ++ F+ FL+ +E G PV+
Sbjct: 373 IVIHDSLM--ASVAQDATNMPNVENYTFHSTCAFTTFLYY--WEVMGRPPVEGFFQATEI 540
Query: 148 PGFEWIHKAQYLLF------------SSIYE----LESQAIDVLK----SKLPLPIYTIG 187
P Q++ F +IY +E I+ L+ SK L + +G
Sbjct: 541 PSMGGCFPPQFIHFITEEYEFHQFNDGNIYNTSRAIEGPYIEFLERIGGSKKRL--WALG 714
Query: 188 PTIPKFSLIKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCA 247
P P ++ K DPK T H IEWLD Q SV+Y++ G+ S + AQ ++IA L
Sbjct: 715 PFNP-LTIEKKDPK---TRH-ICIEWLDKQEANSVMYVSFGTTTSFTVAQFEQIAIGLEQ 879
Query: 248 SNVRFLWIAR----------SEASRLKEICGAHHMG-----LIMEWCDQLRVLSHPSIGG 292
S +F+W+ R SEA R + G LI +W QL +LSH S GG
Sbjct: 880 SKQKFIWVLRDADKGNIFDGSEAERYELPNGFEERVEGIGLLIRDWAPQLEILSHTSTGG 1059
Query: 293 FWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDK 352
F SHCGWNS ES+ GVP P++ DQP NS ++ E KVG VK+ +R+ LV
Sbjct: 1060FMSHCGWNSCLESITMGVPIAAWPMHSDQPRNSVLITEVLKVGFVVKDWAQRNALVSASV 1239
Query: 353 IVKLVHEFMDL-DGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
+ V M+ +G+ ++R+R+ +L+ S GG + + +FI+ +
Sbjct: 1240VENAVRRLMETKEGD---EMRDRAVRLKNCIHRSKYGGGVSRMEMGSFIAHI 1386
>TC216848 similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8, partial
(39%)
Length = 1113
Score = 138 bits (347), Expect = 5e-33
Identities = 87/266 (32%), Positives = 139/266 (51%), Gaps = 12/266 (4%)
Frame = +1
Query: 151 EWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHSYY 210
E + A + F++ +ELE AI+ L S P +Y+IGP + PS ++ +
Sbjct: 109 EKVPSASAVAFNTFHELERDAINALPSMFP-SLYSIGPFPSFLDQSPHKQVPSLGSNLWK 285
Query: 211 -----IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR-------S 258
++WL+S+ SV+Y+ GS +S+ Q+ E A L S FLWI R S
Sbjct: 286 EDTGCLDWLESKEPRSVVYVNFGSITVMSAEQLLEFAWGLANSKKPFLWIIRPDLVIGGS 465
Query: 259 EASRLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIY 318
+ + LI WC Q +VL+HPSIG F +HCGWNST ES+ AGVP L P +
Sbjct: 466 VILSSEFVNETRDRSLIASWCPQEQVLNHPSIGVFLTHCGWNSTTESICAGVPMLCWPFF 645
Query: 319 IDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKL 378
DQP N + + +W++G + DT K++++ KLV+E M GE + + +++ +L
Sbjct: 646 ADQPTNCRYICNEWEIGMEI------DTNAKREELEKLVNELM--VGEKGKKMGQKTMEL 801
Query: 379 QKICLNSIANGGSAHTDFNAFISDVM 404
+K GG ++ + + I +V+
Sbjct: 802 KKKAEEETRPGGGSYMNLDKLIKEVL 879
>TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, partial (46%)
Length = 993
Score = 138 bits (347), Expect = 5e-33
Identities = 79/221 (35%), Positives = 126/221 (56%), Gaps = 19/221 (8%)
Frame = +1
Query: 203 SNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASR 262
++ N + ++WLD Q SV+Y+ GS ++ +Q+ E+A AL + F+W+ R E S+
Sbjct: 226 ASINEHHCLKWLDLQKSKSVVYVCFGSLCNLIPSQLVELALALEDTKRPFVWVIR-EGSK 402
Query: 263 LKEICG----------AHHMGLIME-WCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVP 311
+E+ GLI+ W Q+ +LSH +IGGF +HCGWNST E + AG+P
Sbjct: 403 YQELEKWISEEGFEERTKGRGLIIRGWAPQVLILSHHAIGGFLTHCGWNSTLEGIGAGLP 582
Query: 312 FLTLPIYIDQPFNSKMMVEDWKVGCRV--------KEDVKRDTLVKKDKIVKLVHEFMDL 363
+T P++ DQ N K++ + K+G V E+ K LVKK+ I + + MD
Sbjct: 583 MITWPLFADQFLNEKLVTKVLKIGVSVGVEVPMKFGEEEKTGVLVKKEDINRAICMVMDD 762
Query: 364 DGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVM 404
DGE +++ RER+ KL ++ ++ NGGS+H D + I D+M
Sbjct: 763 DGEESKERRERATKLSEMAKRAVENGGSSHLDLSLLIQDIM 885
>TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferase like
protein, partial (27%)
Length = 820
Score = 136 bits (342), Expect = 2e-32
Identities = 73/197 (37%), Positives = 117/197 (59%), Gaps = 3/197 (1%)
Frame = +2
Query: 210 YIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASRLKEICGA 269
Y+EWLDSQP SV+Y++ G+ ++ Q+ E+A AL S FLW+ R + +++ C
Sbjct: 14 YVEWLDSQPELSVVYVSFGTLAVLADRQMKELARALLDSGYLFLWVIR-DMQGIEDNCRE 190
Query: 270 H--HMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKM 327
G I++WC Q+ VLSH S+G F +HCGWNST ESL +GVP + P + DQ N+KM
Sbjct: 191 ELEQRGKIVKWCSQVEVLSHGSLGCFVTHCGWNSTMESLGSGVPMVAFPQWTDQGTNAKM 370
Query: 328 MVEDWKVGCRVKEDVK-RDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSI 386
+ + WK G RV + V + +V+ ++I K + M G+ ++ R + K + + ++
Sbjct: 371 VQDVWKTGVRVDDKVNVEEGIVEAEEIRKCLDVVMGSGGK-GQEFRRNADKWKCLAREAV 547
Query: 387 ANGGSAHTDFNAFISDV 403
GGS+ ++ F+ DV
Sbjct: 548 TEGGSSDSNMRTFLHDV 598
>TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8,
partial (47%)
Length = 1125
Score = 135 bits (339), Expect = 4e-32
Identities = 88/268 (32%), Positives = 136/268 (49%), Gaps = 16/268 (5%)
Frame = +2
Query: 153 IHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSN-------- 204
I + +LF++ LES ++ L S P +Y IGP F L+ N S+
Sbjct: 152 IQRNTTILFNTFDGLESDVMNALSSMFP-SLYPIGP----FPLLLNQSPQSHLTSLGSNL 316
Query: 205 -TNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR------ 257
+EWL+S+ SV+Y+ GS +S+ Q+ E A L S FLWI R
Sbjct: 317 WNEDLECLEWLESKESRSVVYVNFGSITVMSAEQLLEFAWGLANSKKPFLWIIRPDLVIG 496
Query: 258 -SEASRLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLP 316
S + + LI WC Q +VL+HPSIG F +HCGWNST ES+ AGVP L P
Sbjct: 497 GSVILSSEFVSETRDRSLIASWCPQEQVLNHPSIGVFLTHCGWNSTTESVCAGVPMLCWP 676
Query: 317 IYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSK 376
+ +QP N + + +W++G + DT K++++ KLV+E M GE + +RE+
Sbjct: 677 FFAEQPTNCRYICNEWEIGMEI------DTSAKREEVEKLVNELM--VGEKGKKMREKVM 832
Query: 377 KLQKICLNSIANGGSAHTDFNAFISDVM 404
+L++ GG ++ + + I +V+
Sbjct: 833 ELKRKAEEVTKPGGCSYMNLDKVIKEVL 916
>TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltransferase ,
partial (50%)
Length = 935
Score = 132 bits (332), Expect = 3e-31
Identities = 72/207 (34%), Positives = 114/207 (54%), Gaps = 7/207 (3%)
Frame = +2
Query: 206 NHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR-----SEA 260
N S I+WLD SV+Y+ GS +S + E A L SN+ FLWI R E+
Sbjct: 74 NDSKCIQWLDQWEPSSVIYVNYGSITVMSEDHLKEFAWGLANSNLPFLWIKRPDLVMGES 253
Query: 261 SRLKE--ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIY 318
++L + + G I WC Q +VLSHPS+G F +HCGWNST E + GVP + P +
Sbjct: 254 TQLPQDFLDEVKDRGYITGWCPQEQVLSHPSVGVFLTHCGWNSTLEGISGGVPMIGWPFF 433
Query: 319 IDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKL 378
+Q N + + W +G +K+DVKR +++ LV E + GE +++R++ +
Sbjct: 434 AEQQTNCRYICTTWGIGMDIKDDVKR------EEVTTLVKEM--ITGERGKEMRQKCLEW 589
Query: 379 QKICLNSIANGGSAHTDFNAFISDVMH 405
+K + + GGS++ DF+ + +V+H
Sbjct: 590 KKKAIEATDMGGSSYNDFHRLVKEVLH 670
>TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (59%)
Length = 1152
Score = 130 bits (326), Expect = 1e-30
Identities = 85/267 (31%), Positives = 138/267 (50%), Gaps = 21/267 (7%)
Frame = +2
Query: 159 LLFSSIYELESQAIDVLKSKLPLPIYTIGPT--IPKFSLIKNDPKPSNTNHSYYIEWLDS 216
++ +S ELE K ++ IGP I K L K ++ + S +I+WLD
Sbjct: 62 VVMNSFEELEPAYATGYKKIRGDKLWCIGPVSLINKDHLDKAQRGTASIDVSQHIKWLDC 241
Query: 217 QPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASR-----------LKE 265
Q G+V+Y GS ++++ Q+ E+ AL AS F+W+ R +E
Sbjct: 242 QKPGTVIYACLGSLCNLTTPQLKELGLALEASKRPFIWVIREGGHSEELEKWIKEYGFEE 421
Query: 266 ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNS 325
A + LI W Q+ +LSHP+IGGF +HCGWNST E++ AGVP LT P++ DQ N
Sbjct: 422 RTNARSL-LIRGWAPQILILSHPAIGGFITHCGWNSTLEAICAGVPMLTWPLFADQFLNE 598
Query: 326 KMMVEDWKVGCRV--------KEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKK 377
++V KVG +V ++V+ VKK + + + + MD E + R+R ++
Sbjct: 599 SLVVHVLKVGVKVGVEIPLTWGKEVEIGVQVKKKDVERAIAKLMDETSE-SEKRRKRVRE 775
Query: 378 LQKICLNSIANGGSAHTDFNAFISDVM 404
L ++ ++ GGS++++ I D+M
Sbjct: 776 LAEMANRAVEKGGSSYSNVTLLIQDIM 856
>BM892443
Length = 422
Score = 127 bits (320), Expect = 7e-30
Identities = 63/105 (60%), Positives = 75/105 (71%)
Frame = +2
Query: 45 KPDNISFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLY 104
KPDNI F + NVIPSE D F+E VMTKMEAPFE+LL+ L PP++I+YDT L+
Sbjct: 2 KPDNIRFATIPNVIPSEHGRANDFVTFVEAVMTKMEAPFEDLLNRL-LPPTVIIYDTYLF 178
Query: 105 WAVVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVKYPG 149
W V VAN+R+IP A FWPM AS F+V H H+ EQNGHYPV G
Sbjct: 179 WVVRVANKRSIPVASFWPMSASFFAVLKHYHLLEQNGHYPVNVSG 313
>TC209756
Length = 1356
Score = 125 bits (315), Expect = 3e-29
Identities = 79/251 (31%), Positives = 129/251 (50%), Gaps = 7/251 (2%)
Frame = +1
Query: 157 QYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSN--TNHSYYIEWL 214
++ L ++ Y+LE A V LP IGP L+++D S + +EWL
Sbjct: 172 EWWLCNTTYDLEPGAFSVSPKFLP-----IGP------LMESDNSKSAFWEEDTTCLEWL 318
Query: 215 DSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASRLKEICGAHH--- 271
D QP SV+Y++ GS + Q E+A AL + F+W+ R + + H
Sbjct: 319 DQQPPQSVIYVSFGSLAVMDPNQFKELALALDLLDKPFIWVVRPCNDNKENVNAYAHDFH 498
Query: 272 --MGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMV 329
G I+ W Q ++L+HP++ F SHCGWNST E + AGVPFL P DQ + +
Sbjct: 499 GSKGKIVGWAPQKKILNHPALASFISHCGWNSTLEGICAGVPFLCWPCATDQYLDKSYIC 678
Query: 330 EDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANG 389
+ WK+G + +D + ++ +++I K V + + + DI+ RS KL+ + +N+I G
Sbjct: 679 DVWKIGLGLDKD--ENGIISREEIRKKVDQLL-----VDEDIKARSLKLKDMTINNILEG 837
Query: 390 GSAHTDFNAFI 400
G + + N F+
Sbjct: 838 GQSSKNLNFFM 870
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 124 bits (311), Expect = 8e-29
Identities = 116/453 (25%), Positives = 194/453 (42%), Gaps = 61/453 (13%)
Frame = +1
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+P P ++ P++ K L+ + H++ + S SS+ + S +P
Sbjct: 16 VPSPGFTHLVPILEFSKRLLHLHPEFHITCFIPSVGSSPTSSKAYVQTLPPTITSIFLPP 195
Query: 61 ELICGRDHPAFME---DVMTKMEAPF--EELLDLLDHPPSI-IVYDTLLYWAVVVANRRN 114
+ P+ + ++ + P+ EEL L + +V D A+ A N
Sbjct: 196 ITLDHVSDPSVLALQIELSVNLSLPYIREELKSLCSRAKVVALVVDVFANGALNFAKELN 375
Query: 115 IPAALFWPMPASIFSVFLHQHIFEQ-------NGHYPVKYPGFEWIHKAQYLL------- 160
+ + ++ P A + S++ + ++ P+ PG IH L
Sbjct: 376 LLSYIYLPQSAMLLSLYFYSTKLDEILSSESRELQKPIDIPGCVPIHNKDLPLPFHDLSG 555
Query: 161 --------------------FSSIYELESQAIDVLKS--KLPLPIYTIGPTIPKFSLIKN 198
++ ELES AI L+ K +Y +GP I S+
Sbjct: 556 LGYKGFLERSKRFHVPDGVFMNTFLELESGAIRALEEHVKGKPKLYPVGPIIQMESI--- 726
Query: 199 DPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARS 258
+ N + WLD Q SVLY++ GS ++S Q +E+A L S +FLW+ R+
Sbjct: 727 ----GHENGVECLTWLDKQEPNSVLYVSFGSGGTLSQEQFNELAFGLELSGKKFLWVVRA 894
Query: 259 EASRLKE--ICG----------------AHHMGLIM-EWCDQLRVLSHPSIGGFWSHCGW 299
+ + +C GL++ W Q++VL H + GGF SHCGW
Sbjct: 895 PSGVVSAGYLCAETKDPLEFLPHGFLERTKKQGLVVPSWAPQIQVLGHSATGGFLSHCGW 1074
Query: 300 NSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHE 359
NS ES+V GVP +T P++ +Q N+ M+ +D KV R K V LV++++I K+V
Sbjct: 1075NSVLESVVQGVPVITWPLFAEQSLNAAMIADDLKVALRPK--VNESGLVEREEIAKVVRG 1248
Query: 360 FMDLDGELTRDIRERSKKLQKICLNSIANGGSA 392
M D E + +IR+R L+ N+I GS+
Sbjct: 1249LMG-DKE-SLEIRKRMGLLKIAAANAIKEDGSS 1341
>TC220859 weakly similar to UP|Q94IF2 (Q94IF2) Glucosyltransferase , partial
(30%)
Length = 1190
Score = 124 bits (310), Expect = 1e-28
Identities = 74/207 (35%), Positives = 114/207 (54%), Gaps = 9/207 (4%)
Frame = +2
Query: 207 HSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASRLKE- 265
H ++WLD QP SV+++ GS S +Q EIA AL S VRFLW S ++ E
Sbjct: 20 HDRILKWLDEQPDSSVVFLCFGSRGSFEPSQTREIALALQHSGVRFLWSMLSPPTKDNEE 199
Query: 266 -------ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIY 318
+ G++ EW Q+ +L+H ++ GF SHCGWNS ES+ GVP LT PIY
Sbjct: 200 RILPEGFLEWTEGRGMLCEWAPQVEILAHKALVGFVSHCGWNSILESMWFGVPILTWPIY 379
Query: 319 IDQPFNSKMMVEDWKVGCRVKEDVKRDT-LVKKDKIVKLVHEFMDLDGELTRDIRERSKK 377
+Q N+ MV ++ + +K D +R + LV +++I K + + MD D + ++ K+
Sbjct: 380 AEQQLNAYRMVREFGLAVELKVDYRRGSDLVMEEEIEKGLKQLMDRDNA----VHKKVKQ 547
Query: 378 LQKICLNSIANGGSAHTDFNAFISDVM 404
++++ +I NGGS+ I DVM
Sbjct: 548 MKEMARKAILNGGSSFISVGELI-DVM 625
>TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (35%)
Length = 1017
Score = 122 bits (305), Expect = 4e-28
Identities = 72/203 (35%), Positives = 112/203 (54%), Gaps = 19/203 (9%)
Frame = +2
Query: 211 IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASRLKE----- 265
+ WLD QP SVLY++ GS ++S QI+E+A+ L S RFLW+ R+ ++ +
Sbjct: 14 LSWLDKQPPCSVLYVSFGSGGTLSQNQINELASGLELSGQRFLWVLRAPSNSVSAAYLEA 193
Query: 266 -------------ICGAHHMGLIM-EWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVP 311
+ GL++ W Q++VL H S+GGF SHCGWNST ES+ GVP
Sbjct: 194 SKEDPLQFLPSGFLERTKEKGLVVASWAPQVQVLGHNSVGGFLSHCGWNSTLESVQEGVP 373
Query: 312 FLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDI 371
+T P++ +Q N+ M+ + KV R K D +V+K++I K++ M DGE +
Sbjct: 374 LITWPLFAEQRMNAVMLTDGLKVALRPK--FNEDGIVEKEEIAKVIKCLM--DGEEGIGM 541
Query: 372 RERSKKLQKICLNSIANGGSAHT 394
RER L+ +++ +G S+ T
Sbjct: 542 RERMGNLKDSAASALKDGSSSQT 610
>TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, partial (57%)
Length = 1322
Score = 121 bits (303), Expect = 6e-28
Identities = 77/259 (29%), Positives = 125/259 (47%), Gaps = 20/259 (7%)
Frame = +3
Query: 159 LLFSSIYELESQAIDVLK--SKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHSYYIEWLDS 216
++ ++ +E +AI L + +P P++ +GP I P + WL+
Sbjct: 339 IIVNTFEAIEEEAIRALSEDATVPPPLFCVGPVISA---------PYGEEDKGCLSWLNL 491
Query: 217 QPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASRLKEICGAHHMGL-- 274
QP SV+ + GS S AQ+ EIA L S RFLW+ R+E + A + L
Sbjct: 492 QPSQSVVLLCFGSMGRFSRAQLKEIAIGLEKSEQRFLWVVRTELGGADD--SAEELSLDE 665
Query: 275 ----------------IMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIY 318
+ +W Q +LSH S+GGF +HCGWNS E++ GVP + P+Y
Sbjct: 666 LLPEGFLERTKEKGMVVRDWAPQAAILSHDSVGGFVTHCGWNSVLEAVCEGVPMVAWPLY 845
Query: 319 IDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKL 378
+Q N +MV++ KV VKE+ +D V ++ V E M+ D ++IR+R K+
Sbjct: 846 AEQKMNRMVMVKEMKVALAVKEN--KDGFVSSTELGDRVRELMESD--KGKEIRQRIFKM 1013
Query: 379 QKICLNSIANGGSAHTDFN 397
+ ++A GG++ +
Sbjct: 1014KMSAAEAMAEGGTSRASLD 1070
>TC231452 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8,
partial (38%)
Length = 835
Score = 120 bits (302), Expect = 8e-28
Identities = 68/192 (35%), Positives = 99/192 (51%), Gaps = 7/192 (3%)
Frame = +3
Query: 221 SVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASRLKEIC-------GAHHMG 273
SV+Y+ GS ++ + E A L S FLWI R + + I G
Sbjct: 12 SVIYVNYGSITVMTEHHLKEFAWGLANSKQHFLWIIRPDVVMGESISLPQEFFDAIKDRG 191
Query: 274 LIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWK 333
I WC Q +VLSHPS+G F +HCGWNST ES+ AGVP + P + +Q N K W
Sbjct: 192 YITSWCVQEKVLSHPSVGAFLTHCGWNSTLESISAGVPMICWPFFAEQQTNCKYACTTWG 371
Query: 334 VGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAH 393
+G + DV+R ++I KLV E M GE +++++S + +K + + GGS++
Sbjct: 372 IGMEINHDVRR------EEIAKLVKEMM--MGEKGMEMKQKSLEWKKKAIRATDVGGSSY 527
Query: 394 TDFNAFISDVMH 405
DF I +V H
Sbjct: 528 NDFYKLIKEVFH 563
>TC230868 weakly similar to UP|Q94AB5 (Q94AB5) AT3g46660/F12A12_180
(Glucosyltransferase-like protein), partial (38%)
Length = 765
Score = 120 bits (300), Expect = 1e-27
Identities = 71/207 (34%), Positives = 110/207 (52%), Gaps = 11/207 (5%)
Frame = +1
Query: 211 IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR------SE----- 259
+ WLD Q SV+Y++ GS ++S A+ EIA L S FLW+ R SE
Sbjct: 10 MSWLDQQDRNSVVYVSFGSIAAISEAEFLEIAWGLANSKQPFLWVIRPGLIHGSEWFEPL 189
Query: 260 ASRLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYI 319
S E G G I++W Q +VLSHP++G FW+H GWNST ES+ GVP + +P +
Sbjct: 190 PSGFLENLGGR--GYIVKWAPQEQVLSHPAVGAFWTHNGWNSTLESICEGVPMICMPCFA 363
Query: 320 DQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQ 379
DQ N+K W+VG +++ + R + K K + + G+ +IRE + L+
Sbjct: 364 DQKVNAKYASSVWRVGVQLQNKLDRGEVEKTIKTLMV--------GDEGNEIRENALNLK 519
Query: 380 KICLNSIANGGSAHTDFNAFISDVMHL 406
+ S+ GGS++ + +SD++ L
Sbjct: 520 EKVNVSLKQGGSSYCFLDRLVSDILSL 600
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,807,494
Number of Sequences: 63676
Number of extensions: 349672
Number of successful extensions: 1946
Number of sequences better than 10.0: 182
Number of HSP's better than 10.0 without gapping: 1858
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1874
length of query: 406
length of database: 12,639,632
effective HSP length: 99
effective length of query: 307
effective length of database: 6,335,708
effective search space: 1945062356
effective search space used: 1945062356
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC141923.7


