

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141863.4 + phase: 0 /partial
(1196 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
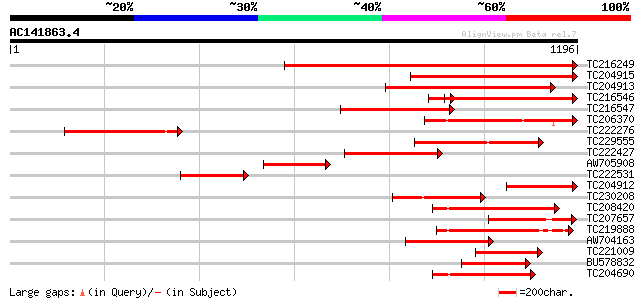
Score E
Sequences producing significant alignments: (bits) Value
TC216249 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 1107 0.0
TC204915 weakly similar to GB|AAH46286.1|28279482|BC046286 spast... 606 e-173
TC204913 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 576 e-164
TC216546 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 511 e-145
TC216547 416 e-116
TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, part... 398 e-111
TC222276 384 e-106
TC229555 similar to GB|AAM62497.1|21553404|AY085265 26S proteaso... 281 9e-76
TC222427 271 2e-72
AW705908 245 7e-65
TC222531 233 4e-61
TC204912 weakly similar to GB|AAH46286.1|28279482|BC046286 spast... 225 8e-59
TC230208 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic ... 211 1e-54
TC208420 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (79%) 209 8e-54
TC207657 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic ... 207 2e-53
TC219888 similar to UP|Q944N4 (Q944N4) Tobacco mosaic virus heli... 207 3e-53
AW704163 201 1e-51
TC221009 homologue to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, pa... 198 1e-50
BU578832 195 1e-49
TC204690 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type A... 181 2e-45
>TC216249 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial (14%)
Length = 2222
Score = 1107 bits (2864), Expect = 0.0
Identities = 551/618 (89%), Positives = 587/618 (94%)
Frame = +2
Query: 579 KVVLIFDDNPLSKIGVRFDKPIPDGVDLGSACEAGQGFFCNITDLRLENSGIDELDKSLI 638
KVVL+FDDNPLSKIGVRFDKPIPDGVDLG ACE GQGFFCN+TDLRLE+S ++ELDK LI
Sbjct: 17 KVVLLFDDNPLSKIGVRFDKPIPDGVDLGGACEGGQGFFCNVTDLRLESSAVEELDKLLI 196
Query: 639 NTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYSFKSKLEKLPDNVVVIGSHTHSDSRK 698
++LFEVV SESR +PFILFMK+AEKSIVGNGD +SFKSKLE LPDNVVVIGSHT +DSRK
Sbjct: 197 HSLFEVVFSESRSAPFILFMKDAEKSIVGNGDSHSFKSKLENLPDNVVVIGSHTQNDSRK 376
Query: 699 EKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQ 758
EKSH GGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKE PK N+TLTKLFPNK+TIHMPQ
Sbjct: 377 EKSHPGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEAPKQNRTLTKLFPNKITIHMPQ 556
Query: 759 DEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKI 818
DEALLASWKQQLDRDVETLKIKGNLHHLRTVL R GME +GLE+LC+KD TLTNEN+EKI
Sbjct: 557 DEALLASWKQQLDRDVETLKIKGNLHHLRTVLGRCGMECEGLETLCIKDQTLTNENAEKI 736
Query: 819 LGWALSHHLMQNPEADADAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEK 878
+GWALSHHLMQN EA D+KLVLS ESI YGIGI Q+IQNESKSLKKSLKDVVTENEFEK
Sbjct: 737 IGWALSHHLMQNSEAKPDSKLVLSCESILYGIGILQSIQNESKSLKKSLKDVVTENEFEK 916
Query: 879 RLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGP 938
RLL DVIPP+DI VTFDDIGALE VKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGP
Sbjct: 917 RLLADVIPPSDIDVTFDDIGALEKVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGP 1096
Query: 939 PGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEV 998
PGTGKTMLAKA+AT+AGANFINISMSSITSKWFGEGEKYVKAVFSLASKI+PSVIFVDEV
Sbjct: 1097 PGTGKTMLAKAIATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKISPSVIFVDEV 1276
Query: 999 DSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPR 1058
DSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTK+TERV+VLAATNRP+DLDEAVIRR+PR
Sbjct: 1277 DSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKETERVLVLAATNRPFDLDEAVIRRMPR 1456
Query: 1059 RLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE 1118
RLMVNLPDAPNRAKILKVILAKE+LS DVDL A+A+MTDGYSGSDLKNLCVTAAHRPIKE
Sbjct: 1457 RLMVNLPDAPNRAKILKVILAKEELSPDVDLDAVASMTDGYSGSDLKNLCVTAAHRPIKE 1636
Query: 1119 ILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQW 1178
ILEKEKKE AAA+AEG+PAPAL S D+RSLNMEDFK+AHQQVCASVSSESVNMTEL+QW
Sbjct: 1637 ILEKEKKERAAALAEGQPAPALCSSGDVRSLNMEDFKYAHQQVCASVSSESVNMTELLQW 1816
Query: 1179 NELYGEGGSRVKKALSYF 1196
NELYGEGGSRVKKALSYF
Sbjct: 1817 NELYGEGGSRVKKALSYF 1870
>TC204915 weakly similar to GB|AAH46286.1|28279482|BC046286 spastic paraplegia
4 homolog {Mus musculus;} , partial (21%)
Length = 1470
Score = 606 bits (1563), Expect = e-173
Identities = 302/352 (85%), Positives = 324/352 (91%)
Frame = +1
Query: 845 SIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVK 904
S+ YGI I Q IQNE+K+LKKSLKDVVTENEFEK+LL DVIPP DIGVTFDDIGALENVK
Sbjct: 130 SLNYGINILQGIQNENKNLKKSLKDVVTENEFEKKLLADVIPPTDIGVTFDDIGALENVK 309
Query: 905 DTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMS 964
DTLKELVMLPLQRPELFCKGQL KPCKGILLFGPPGTGKTMLAKAVAT+AGANFINISMS
Sbjct: 310 DTLKELVMLPLQRPELFCKGQLAKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMS 489
Query: 965 SITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVN 1024
SITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENP EHEAMRKMKNEFMVN
Sbjct: 490 SITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPSEHEAMRKMKNEFMVN 669
Query: 1025 WDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS 1084
WDGLRTKD ERV+VLAATNRP+DLDEAVIRRLPRRLMVNLPDAPNR KIL+VIL KEDL+
Sbjct: 670 WDGLRTKDKERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDAPNREKILRVILVKEDLA 849
Query: 1085 SDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSD 1144
DVD AIANMTDGYSGSDLKNLCVTAAH PI+EILEKEKKE + A++E +P P L GS
Sbjct: 850 PDVDFEAIANMTDGYSGSDLKNLCVTAAHCPIREILEKEKKERSLALSESKPLPGLCGSG 1029
Query: 1145 DIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
DIR L M+DF++AH+QVCASVSSES NM EL+QWN+LYGEGGSR ++LSYF
Sbjct: 1030 DIRPLKMDDFRYAHEQVCASVSSESTNMNELLQWNDLYGEGGSRKMRSLSYF 1185
>TC204913 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial (14%)
Length = 1080
Score = 576 bits (1484), Expect = e-164
Identities = 291/359 (81%), Positives = 322/359 (89%), Gaps = 1/359 (0%)
Frame = +3
Query: 794 GMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQNPEADA-DAKLVLSSESIQYGIGI 852
G++ LE+LC+KD TLT E+ EKI+GWA+S+H M + EA D+KLV+S+ESI+YG I
Sbjct: 3 GLDCPDLETLCIKDHTLTTESVEKIIGWAISYHFMHSSEASIRDSKLVISAESIKYGHKI 182
Query: 853 FQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVM 912
Q IQNE+K++KKSLKDVVTENEFEK+LL DVIPP DIGVTFDDIGALENVK+TLKELVM
Sbjct: 183 LQGIQNENKNMKKSLKDVVTENEFEKKLLTDVIPPTDIGVTFDDIGALENVKETLKELVM 362
Query: 913 LPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFG 972
LPLQRPELF KGQL KPCKGILLFGPPGTGKTMLAKAVAT+AGANFINISMSSITSKWFG
Sbjct: 363 LPLQRPELFGKGQLAKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFG 542
Query: 973 EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD 1032
EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD
Sbjct: 543 EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD 722
Query: 1033 TERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAI 1092
ER++VLAATNRP+DLDEAVIRRLPRRLMVNLPDAPNR KI++VILAKEDL+ DVD AI
Sbjct: 723 KERILVLAATNRPFDLDEAVIRRLPRRLMVNLPDAPNRGKIVRVILAKEDLAPDVDFEAI 902
Query: 1093 ANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNM 1151
ANMTDGYSGSDLKNLCVTAA PI++ILEKEKKE + A+AE +P P L S D+R L M
Sbjct: 903 ANMTDGYSGSDLKNLCVTAAQCPIRQILEKEKKERSLALAENQPLPQLCSSTDVRPLKM 1079
>TC216546 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial (14%)
Length = 1321
Score = 511 bits (1317), Expect = e-145
Identities = 257/279 (92%), Positives = 266/279 (95%)
Frame = +3
Query: 918 PELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKY 977
P F KG PCKGILLFGPPGTGKTMLAKAVAT+AGANFINISMSSITSKWFGEGEKY
Sbjct: 105 PSYFAKGN*PXPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKY 284
Query: 978 VKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVI 1037
VKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERV+
Sbjct: 285 VKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVL 464
Query: 1038 VLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTD 1097
VLAATNRP+DLDEAVIRRLPRRLMVNLPDAPNRAKILKVIL KEDLSSD+D+ AIA+MTD
Sbjct: 465 VLAATNRPFDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILEKEDLSSDIDMDAIASMTD 644
Query: 1098 GYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHA 1157
GYSGSDLKNLCVTAAHRPIKEILEKEKKE AAAV+EGRPAPAL GS DIRSLNMEDFK+A
Sbjct: 645 GYSGSDLKNLCVTAAHRPIKEILEKEKKEQAAAVSEGRPAPALSGSGDIRSLNMEDFKYA 824
Query: 1158 HQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
HQQVCASVSSES+NMTEL QWNELYGEGGSRVKKALSYF
Sbjct: 825 HQQVCASVSSESINMTELQQWNELYGEGGSRVKKALSYF 941
Score = 96.7 bits (239), Expect = 6e-20
Identities = 47/56 (83%), Positives = 49/56 (86%)
Frame = +2
Query: 884 VIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPP 939
VIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTK +G + P
Sbjct: 2 VIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKXVQGYPIIWSP 169
>TC216547
Length = 733
Score = 416 bits (1069), Expect = e-116
Identities = 207/240 (86%), Positives = 220/240 (91%)
Frame = +2
Query: 699 EKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQ 758
EKSH GGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKE PKPNKTLTKLFPNKVTIHMPQ
Sbjct: 2 EKSHPGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEAPKPNKTLTKLFPNKVTIHMPQ 181
Query: 759 DEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKI 818
DEALLASWKQQLDRDVETLKIK NLH+LRTVLSR G+E +GLE+LC+++ TL+ EN+EKI
Sbjct: 182 DEALLASWKQQLDRDVETLKIKENLHNLRTVLSRCGVECEGLETLCIRNQTLSIENAEKI 361
Query: 819 LGWALSHHLMQNPEADADAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEK 878
+GWALS HLMQN E D DAKLVLS +SIQYG+GI A QNESKSLKKSLKDVVTENEFEK
Sbjct: 362 VGWALSCHLMQNAETDPDAKLVLSCKSIQYGVGILHATQNESKSLKKSLKDVVTENEFEK 541
Query: 879 RLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGP 938
RLL DVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQL +PC GILL P
Sbjct: 542 RLLADVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLXQPCXGILLLVP 721
>TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (38%)
Length = 1502
Score = 398 bits (1022), Expect = e-111
Identities = 201/332 (60%), Positives = 258/332 (77%), Gaps = 10/332 (3%)
Frame = +2
Query: 875 EFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGIL 934
EFEKR+ +VIP N+IGVTF DIGAL+ +K++L+ELVMLPL+RP+LF KG L KPC+GIL
Sbjct: 2 EFEKRIRPEVIPANEIGVTFADIGALDEIKESLQELVMLPLRRPDLF-KGGLLKPCRGIL 178
Query: 935 LFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIF 994
LFGPPGTGKTMLAKA+A +AGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K+AP++IF
Sbjct: 179 LFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVAPTIIF 358
Query: 995 VDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR 1054
VDEVDSMLG+R GEHEAMRK+KNEFM +WDGL T E+++VLAATNRP+DLDEA+IR
Sbjct: 359 VDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLLTGPNEQILVLAATNRPFDLDEAIIR 538
Query: 1055 RLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHR 1114
R RR++V LP NR ILK +LAKE ++D +A MT+GY+GSDLKNLC+TAA+R
Sbjct: 539 RFERRILVGLPSVENREMILKTLLAKEK-HENLDFKELATMTEGYTGSDLKNLCITAAYR 715
Query: 1115 PIKEILEKEK---KELAAAVAEGRPAPALRGSDD-------IRSLNMEDFKHAHQQVCAS 1164
P++E++++E+ E AEG+ + + D +R LNMED + A QV AS
Sbjct: 716 PVRELIQQERMKDMEKKKREAEGQSSEDASNNKDKEEKEITLRPLNMEDMRQAKTQVAAS 895
Query: 1165 VSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
+SE M EL WN+LYGEGGSR K+ L+YF
Sbjct: 896 FASEGSVMNELKHWNDLYGEGGSRKKQQLTYF 991
>TC222276
Length = 748
Score = 384 bits (986), Expect = e-106
Identities = 195/250 (78%), Positives = 212/250 (84%), Gaps = 1/250 (0%)
Frame = +2
Query: 116 NFHLKDHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIVNGTLVKKSTCCTLNSGDEVV 175
NF LKD TIS NLCKIKHTQREG+ VAVLES GSKGSV+VNGTLVK+S C LNSGDEVV
Sbjct: 2 NFSLKDQTISANLCKIKHTQREGNVVAVLESMGSKGSVVVNGTLVKRSASCVLNSGDEVV 181
Query: 176 FGLHGNHSYIFQQVNTEVAVKGAEVQSGIGKFMQLERRSGDPSAVAGASILASLSNLRQD 235
FGL GNHSYIFQQ+N EV VK AE+Q G GKF Q ERR+GDPSAVAGASILASLS+LR +
Sbjct: 182 FGLLGNHSYIFQQINPEVTVKAAEIQGGAGKFFQFERRAGDPSAVAGASILASLSSLRPE 361
Query: 236 LTRWKSPSQTASKPHQGADVSIHTVLPDGTEIELDGL-GNSTPSMGTDKAADAEASNKNT 294
LTRWKSPSQTA+KP QG DVS H+VLPDGTE ELDGL GNS P++ TDKA+D S+KN+
Sbjct: 362 LTRWKSPSQTANKPQQGTDVSSHSVLPDGTETELDGLEGNSAPNVATDKASDVGTSDKNS 541
Query: 295 PMDCDPEDAGAEPGNVKYSGVNDLLRPFFRILAGSTTCKLKLSKSICKQVLEERNGAEDT 354
PMDCDP+DAG E GNVK SGVN L PFFR+LAGS TCKLKLSKSICKQV EERNG D
Sbjct: 542 PMDCDPDDAGTEAGNVKISGVNAFLGPFFRVLAGS-TCKLKLSKSICKQVFEERNGTRDA 718
Query: 355 QAASTSGTSV 364
QAASTSGTSV
Sbjct: 719 QAASTSGTSV 748
>TC229555 similar to GB|AAM62497.1|21553404|AY085265 26S proteasome regulatory
particle chain RPT6-like protein {Arabidopsis thaliana;}
, partial (85%)
Length = 1155
Score = 281 bits (720), Expect = 9e-76
Identities = 138/272 (50%), Positives = 197/272 (71%)
Frame = +2
Query: 854 QAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVML 913
+A++++ + K+ + ++ N +E + D+I P+ I V F+ IG LE +K L ELV+L
Sbjct: 119 KALEHKKEIAKRLGRPLIQTNPYEDVIACDIINPDHIDVEFNSIGGLETIKQALFELVIL 298
Query: 914 PLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGE 973
PL+RP+LF G+L P KG+LL+GPPGTGKTMLAKA+A ++ A FIN+ +S++ SKWFG+
Sbjct: 299 PLKRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESRAVFINVRISNLMSKWFGD 478
Query: 974 GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDT 1033
+K V AVFSLA K+ P++IF+DEVDS LG+R +HEAM MK EFM WDG T
Sbjct: 479 AQKLVAAVFSLAYKLQPAIIFIDEVDSFLGKRRGT-DHEAMLNMKTEFMALWDGFTTDQN 655
Query: 1034 ERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIA 1093
+V+VLAATNRP +LDEA++RRLP+ + +PD RA+ILKV+L E + ++D G IA
Sbjct: 656 AQVMVLAATNRPSELDEAILRRLPQAFEIGIPDQRERAEILKVVLKGERVEDNIDFGHIA 835
Query: 1094 NMTDGYSGSDLKNLCVTAAHRPIKEILEKEKK 1125
+ +GY+GSDL +LC AA+ PI+E+L++EKK
Sbjct: 836 GLCEGYTGSDLFDLCKKAAYFPIRELLDEEKK 931
>TC222427
Length = 648
Score = 271 bits (692), Expect = 2e-72
Identities = 134/208 (64%), Positives = 171/208 (81%), Gaps = 1/208 (0%)
Frame = +1
Query: 707 LFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEALLASW 766
+FTKFGSNQTALLDLAFPD+FGRLHDR KE PK K L +LFPNKVTI +PQDEA+L+ W
Sbjct: 25 MFTKFGSNQTALLDLAFPDNFGRLHDRSKETPKVMKQLGRLFPNKVTIQLPQDEAILSDW 204
Query: 767 KQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKILGWALSHH 826
KQQL+RD+ET+K + N+ +RTVL+R G++ LE+L +KD TLT E+ EKI+GWA+S+H
Sbjct: 205 KQQLERDIETMKAQSNIVSIRTVLNRIGLDCPDLETLSIKDQTLTTESVEKIIGWAISYH 384
Query: 827 LMQNPEAD-ADAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVI 885
M + +AD D+KLV+S+ES+ YGI I Q IQ+E+K+ KKSLKDVVT+NE +K+L+ DVI
Sbjct: 385 FMHSSKADIKDSKLVISAESLNYGINILQGIQSENKN*KKSLKDVVTDNECKKKLVADVI 564
Query: 886 PPNDIGVTFDDIGALENVKDTLKELVML 913
PP DI VT DD+ A E+VKDTL+ELVML
Sbjct: 565 PPTDIEVTVDDMRA*EDVKDTLEELVML 648
>AW705908
Length = 425
Score = 245 bits (626), Expect = 7e-65
Identities = 119/141 (84%), Positives = 129/141 (91%)
Frame = +2
Query: 536 STSGTAKNCLFKLGDRVKYSSSSACLYQTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVR 595
STSGTAKNC+FKLGDRVKYSSSS LYQ + +GP+NGSRGKVVL+FDDNPLSKIGVR
Sbjct: 2 STSGTAKNCVFKLGDRVKYSSSSGGLYQLQTISSRGPANGSRGKVVLLFDDNPLSKIGVR 181
Query: 596 FDKPIPDGVDLGSACEAGQGFFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFI 655
FDKPIPDGVDLG CE GQGFFCN+TDLRLENSGI+ELDK LINTLFEVV SESRD+PFI
Sbjct: 182 FDKPIPDGVDLGGLCEPGQGFFCNVTDLRLENSGIEELDKLLINTLFEVVVSESRDAPFI 361
Query: 656 LFMKEAEKSIVGNGDPYSFKS 676
LFMK+AEKSIVGNGDP+SFKS
Sbjct: 362 LFMKDAEKSIVGNGDPFSFKS 424
>TC222531
Length = 429
Score = 233 bits (594), Expect = 4e-61
Identities = 119/144 (82%), Positives = 126/144 (86%)
Frame = +1
Query: 360 SGTSVRCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAKYT 419
+GTSVRCAVFKED HAAILDGKE EVS DNFPYYLSENTKNVLIAAC IHLKHKE KYT
Sbjct: 1 AGTSVRCAVFKEDVHAAILDGKEIEVSLDNFPYYLSENTKNVLIAACVIHLKHKELVKYT 180
Query: 420 ADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDG 479
DL T+NPRILLSGPAGSEIY EML KALAKYFGAKLLIFDS LLGGLSSKEAELLKDG
Sbjct: 181 TDLTTINPRILLSGPAGSEIYQEMLAKALAKYFGAKLLIFDSHSLLGGLSSKEAELLKDG 360
Query: 480 FNAEKSCSCPKQSPTATDMAKSTD 503
FNAEK C+ K SP+++DMA+ D
Sbjct: 361 FNAEKFCAYAK-SPSSSDMARCMD 429
>TC204912 weakly similar to GB|AAH46286.1|28279482|BC046286 spastic paraplegia
4 homolog {Mus musculus;} , partial (4%)
Length = 911
Score = 225 bits (574), Expect = 8e-59
Identities = 113/149 (75%), Positives = 126/149 (83%)
Frame = +2
Query: 1048 LDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNL 1107
LDEAVIRRLPRRLMVNLPDAPNR KI+ VILAKE+L+ DVD AIANMTDGYSGSDLKNL
Sbjct: 14 LDEAVIRRLPRRLMVNLPDAPNREKIVSVILAKEELAPDVDFEAIANMTDGYSGSDLKNL 193
Query: 1108 CVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSS 1167
CVTAAH PI+EILEKEKKE + A+ E +P P L S DIR L MEDF +AH+QVC SVSS
Sbjct: 194 CVTAAHCPIREILEKEKKERSLALTENQPLPQLCSSTDIRPLKMEDFIYAHEQVCVSVSS 373
Query: 1168 ESVNMTELVQWNELYGEGGSRVKKALSYF 1196
ES NM EL+QWN+LYGEGGSR ++LSYF
Sbjct: 374 ESTNMNELLQWNDLYGEGGSRKMRSLSYF 460
>TC230208 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic reticulum
ATPase-like, partial (45%)
Length = 642
Score = 211 bits (538), Expect = 1e-54
Identities = 105/197 (53%), Positives = 146/197 (73%), Gaps = 2/197 (1%)
Frame = +1
Query: 808 LTLTNENSEKILGWALSHHLMQN--PEADADAKLVLSSESIQYGIGIFQAIQNESKSLKK 865
+ LT +EK++GWA +H+L P + +L L ES++ + + + S+ +
Sbjct: 58 IILTKHKAEKVVGWAKNHYLSSCLLPSIKGE-RLYLPRESLEIAVSRLKGQETMSRKPSQ 234
Query: 866 SLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQ 925
SLK++ ++EFE + V+PP +IGV FDDIGALE+VK L ELV+LP++RPELF +G
Sbjct: 235 SLKNLA-KDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFSRGN 411
Query: 926 LTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLA 985
L +PCKGILLFGPPGTGKT+LAKA+AT+AGANFI+I+ S++TSKWFG+ EK KA+FS A
Sbjct: 412 LLRPCKGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSFA 591
Query: 986 SKIAPSVIFVDEVDSML 1002
SK+AP ++FVDEVDS+L
Sbjct: 592 SKLAPVIVFVDEVDSLL 642
>TC208420 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (79%)
Length = 1523
Score = 209 bits (531), Expect = 8e-54
Identities = 117/273 (42%), Positives = 168/273 (60%), Gaps = 5/273 (1%)
Frame = +1
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V ++ I LEN K LKE V++P++ P+ F L P KGILLFGPPGTGKTMLAKAVA
Sbjct: 454 VKWESIKGLENAKRLLKEAVVMPIKYPKYFTG--LLSPWKGILLFGPPGTGKTMLAKAVA 627
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRR-ENPGE 1010
T+ F NIS SS+ SKW G+ EK VK +F LA APS IF+DE+D+++ +R E E
Sbjct: 628 TECKTTFFNISASSVVSKWRGDSEKLVKVLFELARHHAPSTIFLDEIDAIISQRGEARSE 807
Query: 1011 HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNR 1070
HEA R++K E ++ DGL TK E V VLAATN P++LD A++RRL +R++V LP+ R
Sbjct: 808 HEASRRLKTELLIQMDGL-TKTDELVFVLAATNLPWELDAAMLRRLEKRILVPLPEPVAR 984
Query: 1071 AKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAA 1130
+ + +L ++ + + + T+GYSGSD++ LC A +P++ ++ + ++
Sbjct: 985 RAMFEELLPQQPDEEPIPYDILVDKTEGYSGSDIRLLCKETAMQPLRRLMSQLEQSQDVV 1164
Query: 1131 VAEGRPAPALRGSDDI----RSLNMEDFKHAHQ 1159
E P S+DI R+ HAH+
Sbjct: 1165 PEEELPKVGPIKSEDIETALRNTRPSAHLHAHK 1263
>TC207657 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic reticulum
ATPase-like, partial (37%)
Length = 919
Score = 207 bits (528), Expect = 2e-53
Identities = 99/186 (53%), Positives = 137/186 (73%)
Frame = +3
Query: 1010 EHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPN 1069
EHEA R+M+NEFM WDGLR+K+ +R+++L ATNRP+DLD+AVIRRLPRR+ V+LPDA N
Sbjct: 27 EHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAEN 206
Query: 1070 RAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAA 1129
R KIL++ LA+E+L+SD +AN+TDGYSGSDLKNLC+ AA+RP++E+LE+EKK +
Sbjct: 207 RMKILRIFLAQENLNSDFQFDKLANLTDGYSGSDLKNLCIAAAYRPVQELLEEEKKGASN 386
Query: 1130 AVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRV 1189
+ +R LN++DF A +V SV+ ++ +M EL +WNE+YGEGGSR
Sbjct: 387 DT-----------TSILRPLNLDDFIQAKSKVGPSVAYDATSMNELRKWNEMYGEGGSRT 533
Query: 1190 KKALSY 1195
K +
Sbjct: 534 KAPFGF 551
>TC219888 similar to UP|Q944N4 (Q944N4) Tobacco mosaic virus helicase
domain-binding protein (Fragment), partial (50%)
Length = 1119
Score = 207 bits (526), Expect = 3e-53
Identities = 120/290 (41%), Positives = 177/290 (60%), Gaps = 1/290 (0%)
Frame = +3
Query: 901 ENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFIN 960
E K L E+V+LP +R +LF L +P +G+LLFGPPG GKTMLAKAVA+++ A F N
Sbjct: 3 EKAKQALMEMVILPTKRRDLFTG--LRRPARGLLLFGPPGNGKTMLAKAVASESQATFFN 176
Query: 961 ISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNE 1020
++ +S+TSKW GE EK V+ +F +A PSVIF+DE+DS++ R E++A R++K+E
Sbjct: 177 VTAASLTSKWVGEAEKLVRTLFMVAISRQPSVIFIDEIDSIMSTR-LANENDASRRLKSE 353
Query: 1021 FMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAK 1080
F++ +DG+ + + VIV+ ATN+P +LD+AV+RRL +R+ + LPD R +LK L
Sbjct: 354 FLIQFDGVTSNPDDIVIVIGATNKPQELDDAVLRRLVKRIYIPLPDENVRKLLLKHKLKG 533
Query: 1081 EDLS-SDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPA 1139
+ S DL + T+GYSGSDL+ LC AA PI +EL A + +
Sbjct: 534 QAFSLPSRDLERLVKETEGYSGSDLQALCEEAAMMPI--------RELGADILTVK---- 677
Query: 1140 LRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRV 1189
++ +R L EDFK A + S++ EL +WNE +G S +
Sbjct: 678 ---ANQVRGLRYEDFKKAMATIRPSLNKS--KWEELERWNEEFGSN*SHI 812
>AW704163
Length = 559
Score = 201 bits (512), Expect = 1e-51
Identities = 101/185 (54%), Positives = 145/185 (77%)
Frame = +2
Query: 836 DAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFD 895
D+KLV+S+ESI YG I Q IQN++K++KKSL+D++T+NEF+K+LL + IPP DI +TF+
Sbjct: 5 DSKLVISAESINYGHKILQGIQNDNKNMKKSLEDLITKNEFDKKLLTNGIPPTDIKITFN 184
Query: 896 DIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAG 955
+I +LEN K+TL++ V+LPL++P+LF K LTKP +GILLF PP T KT+L K+V T+A
Sbjct: 185 NIESLENFKETLEKWVILPLRKPKLFNKKHLTKPYRGILLFNPPNTKKTILTKSVTTEAK 364
Query: 956 ANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMR 1015
N INIS+SSITSK + EKYVK VFSL +I PS+I ++E +++L +NP HE++R
Sbjct: 365 PNSINISVSSITSK*CSKEEKYVKXVFSLTIEITPSMIVINESNNILQGHQNPT*HESIR 544
Query: 1016 KMKNE 1020
++ N+
Sbjct: 545 EVNNQ 559
>TC221009 homologue to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (18%)
Length = 696
Score = 198 bits (504), Expect = 1e-50
Identities = 90/143 (62%), Positives = 121/143 (83%)
Frame = +1
Query: 982 FSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAA 1041
F+LA+K++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM +WDGL TK ER++VLAA
Sbjct: 1 FTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLLTKQGERILVLAA 180
Query: 1042 TNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSG 1101
TNRP+DLDEA+IRR RR+MV LP NR KIL+ +LAKE + ++++ IA MT+GY+G
Sbjct: 181 TNRPFDLDEAIIRRFERRIMVGLPSVENREKILRTLLAKEKVDNELEFKEIATMTEGYTG 360
Query: 1102 SDLKNLCVTAAHRPIKEILEKEK 1124
SDLKNLC TAA+RP++E++++E+
Sbjct: 361 SDLKNLCTTAAYRPVRELIQQER 429
>BU578832
Length = 437
Score = 195 bits (495), Expect = 1e-49
Identities = 93/146 (63%), Positives = 120/146 (81%)
Frame = +1
Query: 953 DAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHE 1012
+AGA+FIN+S+S+ITSKWFGE EK V+A+FSLA+K+AP++IF+DEVDSMLG+R GEHE
Sbjct: 1 EAGASFINVSISNITSKWFGEDEKNVRALFSLAAKVAPTIIFIDEVDSMLGKRTKYGEHE 180
Query: 1013 AMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAK 1072
AMRK+KNEFM +WDG+ TK ER++VLAATNRP+DLDEA+IRR RR+MV LP A NR
Sbjct: 181 AMRKIKNEFMAHWDGILTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENREM 360
Query: 1073 ILKVILAKEDLSSDVDLGAIANMTDG 1098
ILK +LAKE +D ++ +T+G
Sbjct: 361 ILKTLLAKEKY-EHIDFNELSTITEG 435
>TC204690 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase, partial
(94%)
Length = 1630
Score = 181 bits (459), Expect = 2e-45
Identities = 96/219 (43%), Positives = 144/219 (64%), Gaps = 1/219 (0%)
Frame = +2
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V ++D+ LE+ K L+E V+LP++ P+ F + +P + LL+GPPGTGK+ LAKAVA
Sbjct: 515 VKWNDVAGLESAKQALQEAVILPVKFPQFFTGKR--RPWRAFLLYGPPGTGKSYLAKAVA 688
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
T+A + F ++S S + SKW GE EK V +F +A + APS+IFVDE+DS+ G+R E
Sbjct: 689 TEADSTFFSVSSSDLVSKWMGESEKLVSNLFQMARESAPSIIFVDEIDSLCGQRGEGNES 868
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRA 1071
EA R++K E +V G+ D ++V+VLAATN PY LD+A+ RR +R+ + LPD R
Sbjct: 869 EASRRIKTELLVQMQGVGHND-QKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDLKARQ 1045
Query: 1072 KILKVILAKEDLS-SDVDLGAIANMTDGYSGSDLKNLCV 1109
+ KV L + ++ D +A T+G+SGSD+ ++CV
Sbjct: 1046 HMFKVHLGDTPHNLAESDFEHLARKTEGFSGSDI-SVCV 1159
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,388,709
Number of Sequences: 63676
Number of extensions: 689888
Number of successful extensions: 3863
Number of sequences better than 10.0: 177
Number of HSP's better than 10.0 without gapping: 3573
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3772
length of query: 1196
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1088
effective length of database: 5,762,624
effective search space: 6269734912
effective search space used: 6269734912
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC141863.4


