

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141862.23 + phase: 0
(295 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
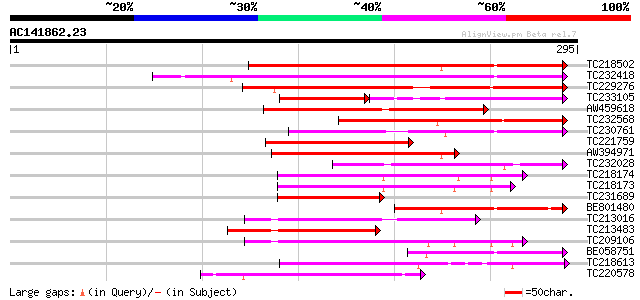
Score E
Sequences producing significant alignments: (bits) Value
TC218502 171 4e-43
TC232418 weakly similar to UP|O81348 (O81348) Symbiotic ammonium... 137 4e-33
TC229276 weakly similar to UP|SRE2_HUMAN (Q12772) Sterol regulat... 125 3e-29
TC233105 68 4e-23
AW459618 101 4e-22
TC232568 weakly similar to UP|Q9ZSQ9 (Q9ZSQ9) Transporter homolo... 97 1e-20
TC230761 weakly similar to UP|O81348 (O81348) Symbiotic ammonium... 91 5e-19
TC221759 homologue to UP|O81348 (O81348) Symbiotic ammonium tran... 91 6e-19
AW394971 85 3e-17
TC232028 84 1e-16
TC218174 similar to UP|Q41102 (Q41102) Phaseolin G-box binding p... 78 4e-15
TC218173 similar to UP|Q41102 (Q41102) Phaseolin G-box binding p... 71 7e-13
TC231689 weakly similar to UP|Q38736 (Q38736) DEL, partial (22%) 70 1e-12
BE801480 66 2e-11
TC213016 homologue to GB|AAN73298.1|25141207|BT002301 At4g16430/... 66 2e-11
TC213483 similar to UP|Q8LPT0 (Q8LPT0) At2g46510/F13A10.4, parti... 64 1e-10
TC209106 similar to UP|Q41101 (Q41101) Phaseolin G-box binding p... 62 2e-10
BE058751 62 3e-10
TC218613 similar to UP|ICE1_ARATH (Q9LSE2) Transcription factor ... 60 9e-10
TC220578 similar to UP|Q93Z71 (Q93Z71) AT5g57150/MUL3_10, partia... 59 2e-09
>TC218502
Length = 873
Score = 171 bits (433), Expect = 4e-43
Identities = 91/171 (53%), Positives = 122/171 (71%), Gaps = 5/171 (2%)
Frame = +3
Query: 125 ENKVSAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQ 184
E K S R+P AQDH+MAER+RREKLSQ FI+L++L+PGLKKMDKA++L DAIK++K+
Sbjct: 24 ETKTSHGKRSPAHAQDHIMAERKRREKLSQSFIALAALVPGLKKMDKASVLGDAIKYVKE 203
Query: 185 LNERVKTLEEHVADKKVESAVFMKRSILFEEDDRSSCDE-----NSDQSLSKIEARVSGK 239
L ER+ LEE + ES V + + L +DD SSCDE + SL ++E+RVSGK
Sbjct: 204 LKERLTVLEEQSKKSRAESVVVLNKPDLSGDDDSSSCDESIGADSVSDSLFEVESRVSGK 383
Query: 240 DMLIRIHGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
+ML+RIH K G +L E++ HHL V +SS+LPFG++ LDITIVAQ+
Sbjct: 384 EMLLRIHCQKQKG-LLVKLLAEIQSHHLFVANSSVLPFGDSILDITIVAQM 533
>TC232418 weakly similar to UP|O81348 (O81348) Symbiotic ammonium
transporter, partial (17%)
Length = 890
Score = 137 bits (346), Expect = 4e-33
Identities = 88/219 (40%), Positives = 127/219 (57%), Gaps = 3/219 (1%)
Frame = +2
Query: 75 PTKLLKASPKIISFDYSNNDSKVKKPKTEIGYGENLNFGS---VISQGDYYKRENKVSAV 131
P +L I+SFD N+ P+T N + + S G +R N+ +
Sbjct: 53 PRRLSSPRTYILSFD--NSTMVPATPETRPRSSNNSPLPAKRALESPGPVARRPNQGAKK 226
Query: 132 NRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKT 191
R Q DHVMAERRRR++L++RFI+LS+ +PGL K DKA++L AI ++KQL ERV+
Sbjct: 227 IRTSSQTIDHVMAERRRRQELTERFIALSATIPGLNKTDKASVLRAAIDYVKQLQERVQE 406
Query: 192 LEEHVADKKVESAVFMKRSILFEEDDRSSCDENSDQSLSKIEARVSGKDMLIRIHGDKHC 251
LE+ + ES +F+K+ D+ ++ + L ++EARV GK++LI IH +K
Sbjct: 407 LEKQDKKRSTESVIFIKKPDPNGNDEDTTST*TNCSILPEMEARVMGKEVLIEIHCEKEN 586
Query: 252 GRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
G IL+ LE HLSV SS+LPFGN+ L ITI Q+
Sbjct: 587 G-VELKILDHLENLHLSVTGSSVLPFGNSALCITITTQM 700
>TC229276 weakly similar to UP|SRE2_HUMAN (Q12772) Sterol regulatory element
binding protein-2 (SREBP-2) (Sterol regulatory
element-binding transcription factor 2), partial (3%)
Length = 1069
Score = 125 bits (313), Expect = 3e-29
Identities = 71/176 (40%), Positives = 108/176 (61%), Gaps = 7/176 (3%)
Frame = +2
Query: 122 YKRENKVSAVNRNPI-------QAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATI 174
+ +E + NR P+ Q DH++AER+RRE +S+ FI+LS+L+P LKKMDKA++
Sbjct: 338 HSKETQEKPHNRKPLKRGRRFSQTLDHILAERKRRENISRMFIALSALIPDLKKMDKASV 517
Query: 175 LEDAIKHLKQLNERVKTLEEHVADKKVESAVFMKRSILFEEDDRSSCDENSDQSLSKIEA 234
L +AI+++K L + VK LE+ +K ES K + +CD+ + K+EA
Sbjct: 518 LSNAIEYVKYLQQHVKDLEQENKKRKTESLGCFKIN--------KTCDDKPIKKCPKVEA 673
Query: 235 RVSGKDMLIRIHGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
RVSGKD+LIR+ +K +L +LE H+L + S++LPFGN+ L IT +A V
Sbjct: 674 RVSGKDVLIRVTCEKQ-KDIVLKLLAKLEAHNLCIVCSNVLPFGNSALSITSIAMV 838
>TC233105
Length = 586
Score = 68.2 bits (165), Expect(2) = 4e-23
Identities = 44/103 (42%), Positives = 60/103 (57%)
Frame = +1
Query: 188 RVKTLEEHVADKKVESAVFMKRSILFEEDDRSSCDENSDQSLSKIEARVSGKDMLIRIHG 247
RV LE+ K ES + +K+S E + C N + L +EARV+ ++LI IH
Sbjct: 187 RVTELEQRKKRGK-ESMIILKKS---EANSEDCCRAN--KMLPDVEARVTENEVLIEIHC 348
Query: 248 DKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
+K G IL+ LE HL V +SS+LPFGN+ L ITI+AQV
Sbjct: 349 EKEDGLELIKILDHLENLHLCVTASSVLPFGNSTLSITIIAQV 477
Score = 57.4 bits (137), Expect(2) = 4e-23
Identities = 25/47 (53%), Positives = 39/47 (82%)
Frame = +3
Query: 141 HVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNE 187
H+MAER+RR++L+Q FI+LS+ +PGL K DK+++L AI ++KQL +
Sbjct: 45 HIMAERKRRQQLTQSFIALSATIPGLNKKDKSSMLGKAIDYVKQLQD 185
>AW459618
Length = 372
Score = 101 bits (252), Expect = 4e-22
Identities = 51/117 (43%), Positives = 80/117 (67%)
Frame = +2
Query: 133 RNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTL 192
R + + H++AER+RR++L+ I+LS+ +PGLKKMDKA +L +A+ + KQL ERVK L
Sbjct: 26 RRSCETRHHIIAERKRRQELTGSIIALSATIPGLKKMDKAYVLREAVNYTKQLQERVKEL 205
Query: 193 EEHVADKKVESAVFMKRSILFEEDDRSSCDENSDQSLSKIEARVSGKDMLIRIHGDK 249
E KV+SA F+++S + + +C+ N + SL ++EARV K++LI IH +K
Sbjct: 206 EN---QNKVDSATFIRKSEASSDKNTGNCETNKEISLFEVEARVLDKEVLIGIHCEK 367
>TC232568 weakly similar to UP|Q9ZSQ9 (Q9ZSQ9) Transporter homolog
(Fragment), partial (17%)
Length = 955
Score = 96.7 bits (239), Expect = 1e-20
Identities = 57/121 (47%), Positives = 82/121 (67%), Gaps = 2/121 (1%)
Frame = +1
Query: 172 ATILEDAIKHLKQLNERVKTLEEHVADKKVESAVFMKRSILFEEDDRSSC--DENSDQSL 229
A++L DAIK++K+L ER++ LEE ++ VES V +K+ L DD S+ DE + + L
Sbjct: 1 ASVLGDAIKYVKELQERMRMLEEEDKNRDVESVVMVKKQRLSCCDDGSASHEDEENSERL 180
Query: 230 SKIEARVSGKDMLIRIHGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQ 289
++EARV KD+L+RIH K G IL E++ HL V +SS+LPFG++ LDITIVAQ
Sbjct: 181 PRVEARVLEKDVLLRIHCQKQKGLLLN-ILVEIQNLHLFVVNSSVLPFGDSVLDITIVAQ 357
Query: 290 V 290
+
Sbjct: 358 M 360
>TC230761 weakly similar to UP|O81348 (O81348) Symbiotic ammonium
transporter, partial (13%)
Length = 697
Score = 91.3 bits (225), Expect = 5e-19
Identities = 57/155 (36%), Positives = 93/155 (59%), Gaps = 10/155 (6%)
Frame = +1
Query: 146 RRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHVADKKVESAV 205
++R+ +L++RF++LS+ +PG K DK +IL +A ++KQL +RV+ LE+ V
Sbjct: 106 KKRKRELAERFLALSATIPGFTKTDKTSILANASSYVKQLQQRVRELEQEV--------- 258
Query: 206 FMKRSILFEEDDRSSCDENS----------DQSLSKIEARVSGKDMLIRIHGDKHCGRTA 255
+ ++ E SSC+ NS ++ L +++ RV KD+LI IH +K G
Sbjct: 259 --QSNVSSNEGATSSCEVNSSNDYYSGGGPNEILPEVKVRVLQKDVLIIIHCEKQKG-IM 429
Query: 256 TAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
IL++LE +LSV +SS+L FG LDITI+A++
Sbjct: 430 LKILSQLENVNLSVVNSSVLRFGKITLDITIIAKM 534
>TC221759 homologue to UP|O81348 (O81348) Symbiotic ammonium transporter,
partial (70%)
Length = 818
Score = 90.9 bits (224), Expect = 6e-19
Identities = 46/78 (58%), Positives = 63/78 (79%), Gaps = 1/78 (1%)
Frame = +1
Query: 134 NPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLE 193
N + + H++AER+RREKLSQRFI+LS+L+PGLKKMDKA++L +AIK+LKQ+ E+V LE
Sbjct: 583 NCLNLKTHIIAERKRREKLSQRFIALSALVPGLKKMDKASVLGEAIKYLKQMQEKVSALE 762
Query: 194 EHVADKK-VESAVFMKRS 210
E K+ VES V +K+S
Sbjct: 763 EEQNRKRTVESVVIVKKS 816
>AW394971
Length = 301
Score = 85.1 bits (209), Expect = 3e-17
Identities = 43/100 (43%), Positives = 73/100 (73%), Gaps = 2/100 (2%)
Frame = +2
Query: 137 QAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHV 196
+A DH+M+ER +R++L+ +FI+L++ +PGLKKMDKA +L +AI ++KQL ER++ LE +
Sbjct: 2 EALDHIMSERNKRQELTSKFIALAAAIPGLKKMDKAHVLREAINYVKQLQERIEDLE*DI 181
Query: 197 ADKKVESAVFMKRSILFEEDDRSSCDE--NSDQSLSKIEA 234
VESA+ + RS L +DD ++ +E +++L++ EA
Sbjct: 182 RKNGVESAITIVRSRLCIDDDSNTDEECYGRNEALAEGEA 301
>TC232028
Length = 805
Score = 83.6 bits (205), Expect = 1e-16
Identities = 52/124 (41%), Positives = 75/124 (59%), Gaps = 2/124 (1%)
Frame = +2
Query: 169 MDKATILEDAIKHLKQLNERVKTLEEHVADKKVESAVFMKRSILFEEDDRSSCDENSDQS 228
MDKA +L +A+ KQL ERVK LE KV+SA F+++S + + +C+ N + S
Sbjct: 266 MDKAYVLREAVNCTKQLQERVKELENQ---NKVDSATFIRKSEASSDKNTGNCETNKEIS 436
Query: 229 LSKIEARVSGKDMLIRIHGDKHCGRTAT--AILNELEKHHLSVQSSSILPFGNNYLDITI 286
L ++EARV K++LI IH +K A+L L HLS SS++LPFG + L I I
Sbjct: 437 LFEVEARVLDKEVLIGIHCEKQKDIVFKIHALLRNL---HLSTTSSTVLPFGTSTLIINI 607
Query: 287 VAQV 290
+AQ+
Sbjct: 608 IAQM 619
>TC218174 similar to UP|Q41102 (Q41102) Phaseolin G-box binding protein PG2
(Fragment), partial (49%)
Length = 1345
Score = 78.2 bits (191), Expect = 4e-15
Identities = 52/147 (35%), Positives = 84/147 (56%), Gaps = 17/147 (11%)
Frame = +3
Query: 140 DHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLE------ 193
+HV AER+RREKL+QRF +L +++P + KMDKA++L DAI ++ +L +++TLE
Sbjct: 432 NHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAISYITELKSKLQTLESDKDGM 611
Query: 194 -EHVADKKVESAVFMKRSILFEEDDRSSCDENSDQSLSKI------EARVSGKDMLIRIH 246
+ + K E + + SSC+ N+ S K+ + ++ G D +IRIH
Sbjct: 612 QKQLEGVKKELEKTTENGSSNHAGNSSSCNNNNKLSNQKLIDVLEMDVKILGWDAMIRIH 791
Query: 247 GDK--HCG-RTATAILN-ELEKHHLSV 269
K H G R TA++ +L+ HH +V
Sbjct: 792 CSKKNHPGARLLTALMELDLDVHHANV 872
>TC218173 similar to UP|Q41102 (Q41102) Phaseolin G-box binding protein PG2
(Fragment), partial (28%)
Length = 867
Score = 70.9 bits (172), Expect = 7e-13
Identities = 44/138 (31%), Positives = 75/138 (53%), Gaps = 14/138 (10%)
Frame = +3
Query: 140 DHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLE--EHVA 197
+HV AER+RREKL+QRF +L +++P + KMD A++L DAI ++ +L +++TLE + V
Sbjct: 30 NHVEAERQRREKLNQRFYALRAVVPNVSKMDXASLLGDAISYITELKSKLQTLESDKDVL 209
Query: 198 DKKVESAVFMKRSILFEEDDRSSCDENSDQSLS----------KIEARVSGKDMLIRIHG 247
K++E + +C+ N++ LS +++ ++ G D +I I
Sbjct: 210 HKQLEGVKKELEKTTYNVSSNHACNNNNNNKLSSNQPALIDLVEMDVKIIGWDAMITITC 389
Query: 248 DK--HCGRTATAILNELE 263
K H T L EL+
Sbjct: 390 SKKNHPAATLMTALMELD 443
>TC231689 weakly similar to UP|Q38736 (Q38736) DEL, partial (22%)
Length = 937
Score = 69.7 bits (169), Expect = 1e-12
Identities = 31/56 (55%), Positives = 45/56 (80%)
Frame = +2
Query: 140 DHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEH 195
+HVM+ERRRR KL+QRF++L S++P + K DK +IL+DAI++LK+L R+ LE H
Sbjct: 707 NHVMSERRRRAKLNQRFLTLRSMVPSISKDDKVSILDDAIEYLKKLERRINELEAH 874
>BE801480
Length = 435
Score = 66.2 bits (160), Expect = 2e-11
Identities = 40/92 (43%), Positives = 64/92 (69%), Gaps = 2/92 (2%)
Frame = +2
Query: 201 VESAVFMKRSILFEEDDRSSCDE--NSDQSLSKIEARVSGKDMLIRIHGDKHCGRTATAI 258
VESA+ + RS L +DD ++ +E +++L ++EARV GK++LI+I+ K G I
Sbjct: 8 VESAITIIRSHLCIDDDSNTDEECYGPNEALPEVEARVLGKEVLIKIYCGKQKG-ILLKI 184
Query: 259 LNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
+++LE+ HL + +S++LPFGN LDITI AQ+
Sbjct: 185 MSQLERLHLYISTSNVLPFGNT-LDITITAQM 277
>TC213016 homologue to GB|AAN73298.1|25141207|BT002301 At4g16430/dl4240w
{Arabidopsis thaliana;} , partial (16%)
Length = 424
Score = 65.9 bits (159), Expect = 2e-11
Identities = 39/123 (31%), Positives = 70/123 (56%)
Frame = +1
Query: 123 KRENKVSAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHL 182
KR K + P+ +HV AER+RREKL+QRF +L +++P + KMDKA++L DAI ++
Sbjct: 7 KRGRKPANGREEPL---NHVEAERQRREKLNQRFYALRAVVPNISKMDKASLLGDAIAYI 177
Query: 183 KQLNERVKTLEEHVADKKVESAVFMKRSILFEEDDRSSCDENSDQSLSKIEARVSGKDML 242
+L ++KT+E E F S+ E + ++ EN ++ +V+ ++
Sbjct: 178 NELQAKLKTIES-------ERERFGSTSMDGPELEANARVENHHNGTPDVDVQVAQDGVI 336
Query: 243 IRI 245
+++
Sbjct: 337 VKV 345
>TC213483 similar to UP|Q8LPT0 (Q8LPT0) At2g46510/F13A10.4, partial (17%)
Length = 914
Score = 63.5 bits (153), Expect = 1e-10
Identities = 34/80 (42%), Positives = 51/80 (63%)
Frame = +2
Query: 114 SVISQGDYYKRENKVSAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKAT 173
SV + KR K + P+ +HV AER+RREKL+QRF +L S++P + KMDKA+
Sbjct: 86 SVADERKPRKRGRKPANGREEPL---NHVEAERQRREKLNQRFYALRSVVPNISKMDKAS 256
Query: 174 ILEDAIKHLKQLNERVKTLE 193
+L D I ++ +L +VK +E
Sbjct: 257 LLGDTIAYINELQAKVKIME 316
>TC209106 similar to UP|Q41101 (Q41101) Phaseolin G-box binding protein PG1,
partial (40%)
Length = 1101
Score = 62.4 bits (150), Expect = 2e-10
Identities = 49/168 (29%), Positives = 82/168 (48%), Gaps = 21/168 (12%)
Frame = +3
Query: 123 KRENKVSAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHL 182
KR K + P+ +HV AER+RREKL+QRF +L +++P + KMDKA++L DAI ++
Sbjct: 267 KRGRKPANGREEPL---NHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAILYI 437
Query: 183 KQLNERVKTLEEHVADKKVESAVFMKRSILFEED---------------DRSSCDENSDQ 227
+L ++ L+ + + + K L ++ S + +
Sbjct: 438 NELKSKLNVLDSEKTELEKQLDSTKKELELATKNPPPPPPPPPPPGPPPSNSVEPKKTTS 617
Query: 228 SLS--KIEARVSGKDMLIRIHGDK--HCGRTATAILN--ELEKHHLSV 269
L+ ++E ++ G D ++RI K H A L +LE HH SV
Sbjct: 618 KLADLELEVKIIGWDAMVRIQCSKKNHPAARLMAALKDLDLEVHHASV 761
>BE058751
Length = 436
Score = 62.0 bits (149), Expect = 3e-10
Identities = 38/92 (41%), Positives = 54/92 (58%), Gaps = 9/92 (9%)
Frame = +1
Query: 208 KRSILFEE---------DDRSSCDENSDQSLSKIEARVSGKDMLIRIHGDKHCGRTATAI 258
K S++F + +D ++ E + L ++E RV GK++LI IH +K G I
Sbjct: 7 KESVIFNKKPDPNGNNNEDTTTSTETNCSILPEMEVRVLGKEVLIEIHCEKENG-VELKI 183
Query: 259 LNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
L+ LE HLSV SS+LPFGN+ L ITI AQ+
Sbjct: 184 LDHLENLHLSVTGSSVLPFGNSSLCITITAQM 279
>TC218613 similar to UP|ICE1_ARATH (Q9LSE2) Transcription factor ICE1
(Inducer of CBF expression 1) (Basic helix-loop-helix
protein 116) (bHLH116) (AtbHLH116), partial (40%)
Length = 1147
Score = 60.5 bits (145), Expect = 9e-10
Identities = 51/167 (30%), Positives = 83/167 (49%), Gaps = 16/167 (9%)
Frame = +3
Query: 141 HVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHVADKK 200
++MAERRRR+KL+ R L S++P + KMD+A+IL DAI++LK+L +R+ L +
Sbjct: 231 NLMAERRRRKKLNDRLYMLRSVVPKISKMDRASILGDAIEYLKELLQRINDLHNELESTP 410
Query: 201 VESAVFMKRSI-------------LFEEDDRSSCDENSDQSLSKIEARVSGKDMLIRIHG 247
V S++ S + EE SS + Q +++E R+ + + IH
Sbjct: 411 VGSSLTPVSSFHPLTPTPPTLPSRIKEELCPSSLPSPNGQP-ARVEVRLR-EGRAVNIH- 581
Query: 248 DKHCGRTATAILN---ELEKHHLSVQSSSILPFGNNYLDITIVAQVK 291
C R + +L+ L+ L +Q + I F +DI Q K
Sbjct: 582 -MFCARKPSLLLSTMRALDNLGLDIQQAVISCFNGFAMDIFRAEQCK 719
>TC220578 similar to UP|Q93Z71 (Q93Z71) AT5g57150/MUL3_10, partial (38%)
Length = 856
Score = 59.3 bits (142), Expect = 2e-09
Identities = 34/121 (28%), Positives = 72/121 (59%), Gaps = 4/121 (3%)
Frame = +1
Query: 100 PKTEIGYGENLNFGS-VISQGD---YYKRENKVSAVNRNPIQAQDHVMAERRRREKLSQR 155
P+T++ + N + GS I G+ +Y + + A ++++ER RR+KL+ R
Sbjct: 283 PETDLFF-PNEDLGSWAIMDGEAVSWYYDSSSPDGTGASSSVASKNIVSERNRRKKLNDR 459
Query: 156 FISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHVADKKVESAVFMKRSILFEE 215
++L +++P + KMDKA+I++DAI++++ L+E+ K ++ + D + + F + F++
Sbjct: 460 LLALRAVVPNITKMDKASIIKDAIEYIQHLHEQEKRIQAEILDLESRNK-FKNPTYEFDQ 636
Query: 216 D 216
D
Sbjct: 637 D 639
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.128 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,826,082
Number of Sequences: 63676
Number of extensions: 127345
Number of successful extensions: 885
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 870
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 873
length of query: 295
length of database: 12,639,632
effective HSP length: 97
effective length of query: 198
effective length of database: 6,463,060
effective search space: 1279685880
effective search space used: 1279685880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC141862.23


