

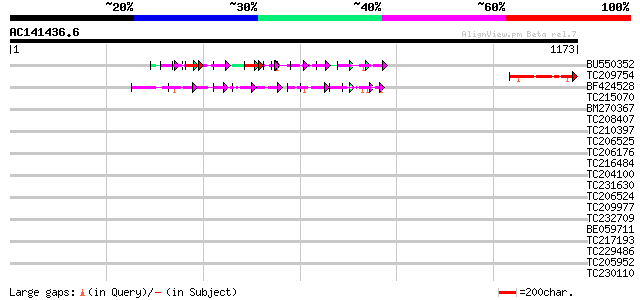
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141436.6 - phase: 0
(1173 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BU550352 114 5e-34
TC209754 133 5e-31
BF424528 127 4e-29
TC215070 similar to UP|Q40336 (Q40336) Proline-rich cell wall pr... 37 0.013
BM270367 similar to PIR|S78452|S784 POU-domain protein rdc-1 - h... 38 0.030
TC208407 similar to UP|Q84KA9 (Q84KA9) RING/C3HC4/PHD zinc finge... 37 0.051
TC210397 37 0.051
TC206525 weakly similar to UP|Q41042 (Q41042) Pisum sativum L. (... 37 0.051
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 37 0.066
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 30 0.076
TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, ... 35 0.15
TC231630 weakly similar to UP|Q7PR65 (Q7PR65) ENSANGP00000016899... 35 0.15
TC206524 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 35 0.15
TC209977 UP|Q9HF30 (Q9HF30) Pheromone MFalpha1D (Fragment), part... 35 0.25
TC232709 weakly similar to UP|Q9LZJ6 (Q9LZJ6) RING-H2 zinc finge... 35 0.25
BE059711 34 0.33
TC217193 similar to UP|Q7XJN2 (Q7XJN2) At2g40650 protein, partia... 34 0.33
TC229486 similar to UP|Q76KV2 (Q76KV2) DNA binding with one fing... 34 0.33
TC205952 34 0.33
TC230110 similar to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP4-... 34 0.43
>BU550352
Length = 531
Score = 114 bits (284), Expect(2) = 5e-34
Identities = 56/146 (38%), Positives = 72/146 (48%), Gaps = 7/146 (4%)
Frame = -1
Query: 526 VECYKTTENQKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKK 585
V CY Q C++ C ++CGRH C +CC D D CS +CG+K
Sbjct: 402 VPCY-----QDLACERKCQRMRDCGRHACKRRCC------------DGDCPPCSEICGRK 274
Query: 586 LRCGQHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGT----MPPLCQLPCSV 641
LRC H C + CH G C PC I ++CACG T +PCGT PP C PC +
Sbjct: 273 LRCKNHKCPSPCHRGPCAPC--PIMVTISCACGETRF--EVPCGTEMDNKPPRCPKPCPI 106
Query: 642 PQPCGHSGS---HSCHFGDCPPCSVP 664
C H+ + H CH+G C PC +P
Sbjct: 105 TPLCRHASNIKPHKCHYGACHPCRLP 28
Score = 80.1 bits (196), Expect(2) = 2e-16
Identities = 48/164 (29%), Positives = 64/164 (38%), Gaps = 4/164 (2%)
Frame = -1
Query: 357 DLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFE 416
DL C ++C ++ DCGRH C+ C G C PC +
Sbjct: 387 DLACERKCQRMRDCGRHACKRRCCDGDCPPCSEI-------------------------- 286
Query: 417 AEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLEDERKSCV 476
CG L C NH C CH G C C + + +C CG+T+ E + +
Sbjct: 285 -----------CGRKLRCKNHKCPSPCHRGPCAPCPIMVT--ISCACGETRFEVPCGTEM 145
Query: 477 D-PIPTCSKVCSKTLRCGVHACK---ETCHVGECPPCKVLISQK 516
D P C K C T C HA CH G C PC++ Q+
Sbjct: 144 DNKPPRCPKPCPITPLCR-HASNIKPHKCHYGACHPCRLPCRQR 16
Score = 77.0 bits (188), Expect(2) = 7e-22
Identities = 38/143 (26%), Positives = 51/143 (35%), Gaps = 6/143 (4%)
Frame = -1
Query: 423 SCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLEDERKSCVDPIPTC 482
+C C + C H C+ C G C P C
Sbjct: 381 ACERKCQRMRDCGRHACKRRCCDGDC-------------------------------PPC 295
Query: 483 SKVCSKTLRCGVHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTENQKFTCQKP 542
S++C + LRC H C CH G C PC ++++ C CG T V C +N+ C KP
Sbjct: 294 SEICGRKLRCKNHKCPSPCHRGPCAPCPIMVTISCACGETRFEVPCGTEMDNKPPRCPKP 115
Query: 543 CGAKKNCGR------HRCSEKCC 559
C C H+C C
Sbjct: 114 CPITPLCRHASNIKPHKCHYGAC 46
Score = 65.1 bits (157), Expect = 2e-10
Identities = 38/103 (36%), Positives = 46/103 (43%)
Frame = -1
Query: 679 IPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQTCGAPRSGCR 738
+PC ++ C C R R CG HAC R C CD G C + CG
Sbjct: 402 VPC-YQDLACERKCQRMRDCGRHACKRRC----CD-------GDCPPCSEICGRKLRCKN 259
Query: 739 HMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGGNN 781
H C + CH G PC A C VTI+C+CG VPC +N
Sbjct: 258 HKCPSPCHRG-PC--APCPIMVTISCACGETRFEVPCGTEMDN 139
Score = 63.5 bits (153), Expect = 5e-10
Identities = 37/122 (30%), Positives = 52/122 (42%), Gaps = 2/122 (1%)
Frame = -1
Query: 338 RLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFC 397
R+ CG+ RC D C + C + L C H C + CH GPC PC +++ SC C
Sbjct: 360 RMRDCGRHACKRRCCDGDCP-PCSEICGRKLRCKNHKCPSPCHRGPCAPCPIMVTISCAC 184
Query: 398 SKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGC--SNHICREVCHPGSCGECEFLP 455
+ + CG E + C C C +++I CH G+C C LP
Sbjct: 183 GETRFEVPCGT-------EMDNKPPRCPKPCPITPLCRHASNIKPHKCHYGACHPCR-LP 28
Query: 456 SR 457
R
Sbjct: 27 CR 22
Score = 59.3 bits (142), Expect(2) = 3e-10
Identities = 43/166 (25%), Positives = 53/166 (31%), Gaps = 7/166 (4%)
Frame = -1
Query: 554 CSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETIFTDL 613
C E+ L P P + C C + CG+H C+ C G CPPC E L
Sbjct: 450 CCEEVMSLWQPQER**VPCYQDLACERKCQRMRDCGRHACKRRCCDGDCPPCSEICGRKL 271
Query: 614 ACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGDCPPCSVPVSKECVGGH 673
C H CH G C PC + V+ C G
Sbjct: 270 RCK-----------------------------NHKCPSPCHRGPCAPCPIMVTISCACGE 178
Query: 674 VILRNIPCG----SNNIKCNNPCGRTRQCGLHACG---RSCHSPPC 712
+PCG + +C PC T C HA CH C
Sbjct: 177 TRF-EVPCGTEMDNKPPRCPKPCPITPLC-RHASNIKPHKCHYGAC 46
Score = 50.4 bits (119), Expect(2) = 5e-34
Identities = 19/40 (47%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Frame = -2
Query: 486 CSKTLRCGVHACKETCHVGEC-PPCKVLISQKCRCGSTSR 524
C K L CG H C E CH G+C C+V++ + CRCGS +
Sbjct: 530 CDKMLPCGYHRCPERCHRGQCVETCRVVVKKSCRCGSLKK 411
Score = 47.8 bits (112), Expect(2) = 2e-09
Identities = 33/117 (28%), Positives = 40/117 (33%), Gaps = 6/117 (5%)
Frame = -1
Query: 635 CQLPCSVPQPCG-HSGSHSCHFGDCPPCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCG 693
C+ C + CG H+ C GDCPPCS CG
Sbjct: 378 CERKCQRMRDCGRHACKRRCCDGDCPPCS---------------------------EICG 280
Query: 694 RTRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQT-----CGAPRSGCRHMCMALC 745
R +C H C CH PC P I+ + ACG+T CG C C
Sbjct: 279 RKLRCKNHKCPSPCHRGPCAPCP-IMVTISCACGETRFEVPCGTEMDNKPPRCPKPC 112
Score = 47.0 bits (110), Expect = 5e-05
Identities = 30/104 (28%), Positives = 37/104 (34%), Gaps = 6/104 (5%)
Frame = -1
Query: 292 CGEPCGKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRC 351
C E CG+ L + H C CH GPC PC C CG+ R C
Sbjct: 297 CSEICGRKLRCK------------NHKCPSPCHRGPCAPCPIMVTIS-CACGETRFEVPC 157
Query: 352 SDRQSDL--TCGQRCDKLLDCGRHHCEN----ACHVGPCDPCQV 389
+ C + C C H N CH G C PC++
Sbjct: 156 GTEMDNKPPRCPKPCPITPLC--RHASNIKPHKCHYGACHPCRL 31
Score = 46.6 bits (109), Expect(2) = 7e-22
Identities = 17/38 (44%), Positives = 24/38 (62%), Gaps = 1/38 (2%)
Frame = -2
Query: 364 CDKLLDCGRHHCENACHVGPC-DPCQVLIEASCFCSKM 400
CDK+L CG H C CH G C + C+V+++ SC C +
Sbjct: 530 CDKMLPCGYHRCPERCHRGQCVETCRVVVKKSCRCGSL 417
Score = 34.3 bits (77), Expect = 0.33
Identities = 17/41 (41%), Positives = 20/41 (48%), Gaps = 1/41 (2%)
Frame = -2
Query: 428 CGNVLGCSNHICREVCHPGSCGE-CEFLPSRVKACCCGKTK 467
C +L C H C E CH G C E C + K+C CG K
Sbjct: 530 CDKMLPCGYHRCPERCHRGQCVETCRVVVK--KSCRCGSLK 414
Score = 33.9 bits (76), Expect(2) = 2e-09
Identities = 16/39 (41%), Positives = 19/39 (48%), Gaps = 2/39 (5%)
Frame = -2
Query: 582 CGKKLRCGQHVCETLCHSGHCPPCLET--IFTDLACACG 618
C K L CG H C CH G C+ET + +C CG
Sbjct: 530 CDKMLPCGYHRCPERCHRGQ---CVETCRVVVKKSCRCG 423
Score = 25.0 bits (53), Expect(2) = 2e-16
Identities = 12/42 (28%), Positives = 18/42 (42%), Gaps = 3/42 (7%)
Frame = -2
Query: 312 DELCP---HACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATR 350
D++ P H C +CH G C + C CG + +R
Sbjct: 527 DKMLPCGYHRCPERCHRGQCVETCRVVVKKSCRCGSLKKDSR 402
Score = 24.6 bits (52), Expect(2) = 3e-10
Identities = 8/16 (50%), Positives = 9/16 (56%)
Frame = -2
Query: 543 CGAKKNCGRHRCSEKC 558
C CG HRC E+C
Sbjct: 530 CDKMLPCGYHRCPERC 483
>TC209754
Length = 587
Score = 133 bits (334), Expect = 5e-31
Identities = 79/151 (52%), Positives = 98/151 (64%), Gaps = 12/151 (7%)
Frame = +3
Query: 1035 QNAGASAASSVTSAWG-------GTKEGALRSNPWKKAAVLDPGWKEDSWGDEQWTTAGD 1087
Q +GAS ASS G G AL+SNPWKK + +PGW+ED+WGDE+W T
Sbjct: 15 QMSGASVASSXYQMPGEDLGQ*KGGALAALKSNPWKKDVIQEPGWREDAWGDEEWATG-- 188
Query: 1088 SANIQPSALKKEAPIPASLNPWNVLNHESSSSSSPATVIRSVASGKQTESGNVSTKVEPS 1147
SAN++ KKEA I AS+NPW+VLN ESSSSSS A I+ S K +ES +V TK+EP
Sbjct: 189 SANVKLPIQKKEARISASVNPWSVLNQESSSSSSVA-AIKIDGSRKHSES-SVITKLEPR 362
Query: 1148 AGGAD-----GGNSDATEAAEVVDDWEKAFE 1173
GG++ GN DA EA++VVDDWEKA E
Sbjct: 363 DGGSNLGGQPAGNFDALEASDVVDDWEKACE 455
>BF424528
Length = 419
Score = 127 bits (318), Expect = 4e-29
Identities = 58/136 (42%), Positives = 71/136 (51%)
Frame = +3
Query: 253 WRCPGCQSVQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKD 312
W CP C+S S Y CFCGK +PP+D ++ PHSCGE CG+PL+
Sbjct: 54 WNCPKCRSEYPKSHIPKTYFCFCGKIENPPNDPWILPHSCGEVCGRPLKHS--------- 206
Query: 313 ELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGR 372
C H C+L CHPGPCP C R C CG RC + + +C + C K LDCG
Sbjct: 207 --CGHHCLLLCHPGPCPSCPKLVKVR-CFCGCLEDVRRCGFK--EFSCNKPCSKFLDCGV 371
Query: 373 HHCENACHVGPCDPCQ 388
H C CH G C PC+
Sbjct: 372 HRCIELCHPGACPPCR 419
Score = 79.7 bits (195), Expect = 7e-15
Identities = 45/91 (49%), Positives = 51/91 (55%), Gaps = 3/91 (3%)
Frame = +3
Query: 423 SCGSNCGNVL--GCSNHICREVCHPGSCGECEFLPSRVKA-CCCGKTKLEDERKSCVDPI 479
SCG CG L C +H C +CHPG C C P VK C CG LED R+ C
Sbjct: 168 SCGEVCGRPLKHSCGHH-CLLLCHPGPCPSC---PKLVKVRCFCGC--LEDVRR-CGFKE 326
Query: 480 PTCSKVCSKTLRCGVHACKETCHVGECPPCK 510
+C+K CSK L CGVH C E CH G CPPC+
Sbjct: 327 FSCNKPCSKFLDCGVHRCIELCHPGACPPCR 419
Score = 79.0 bits (193), Expect = 1e-14
Identities = 47/133 (35%), Positives = 58/133 (43%), Gaps = 9/133 (6%)
Frame = +3
Query: 328 CPPCKAFAPPR------LCPCGKKRIATRCSDRQS-DLTCGQRCDKLL--DCGRHHCENA 378
CP C++ P C CGK I +D +CG+ C + L CG HHC
Sbjct: 60 CPKCRSEYPKSHIPKTYFCFCGK--IENPPNDPWILPHSCGEVCGRPLKHSCG-HHCLLL 230
Query: 379 CHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHI 438
CH GPC C L++ CFC + V CG FSC C L C H
Sbjct: 231 CHPGPCPSCPKLVKVRCFCGCLEDVRRCGFKE-----------FSCNKPCSKFLDCGVHR 377
Query: 439 CREVCHPGSCGEC 451
C E+CHPG+C C
Sbjct: 378 CIELCHPGACPPC 416
Score = 68.9 bits (167), Expect = 1e-11
Identities = 40/109 (36%), Positives = 53/109 (47%), Gaps = 5/109 (4%)
Frame = +3
Query: 461 CCCGKTKLEDERKSCVDP--IP-TCSKVCSKTLR--CGVHACKETCHVGECPPCKVLISQ 515
C CGK + DP +P +C +VC + L+ CG H C CH G CP C L+
Sbjct: 114 CFCGKIENPPN-----DPWILPHSCGEVCGRPLKHSCG-HHCLLLCHPGPCPSCPKLVKV 275
Query: 516 KCRCGSTSRTVECYKTTENQKFTCQKPCGAKKNCGRHRCSEKCCPLSGP 564
+C CG C ++F+C KPC +CG HRC E C P + P
Sbjct: 276 RCFCGCLEDVRRC----GFKEFSCNKPCSKFLDCGVHRCIELCHPGACP 410
Score = 65.5 bits (158), Expect = 1e-10
Identities = 42/123 (34%), Positives = 48/123 (38%), Gaps = 11/123 (8%)
Frame = +3
Query: 601 HCPPCLET-----IFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVP--QPCGHSGSHSC 653
+CP C I C CG PP P +P C C P CGH C
Sbjct: 57 NCPKCRSEYPKSHIPKTYFCFCGKIENPPNDPW-ILPHSCGEVCGRPLKHSCGHHCLLLC 233
Query: 654 HFGDCPPCSVPVSKECVGGHVILRNIP-CGSNNIKCNNPCGRTRQCGLHACGRSCH---S 709
H G CP C V C G L ++ CG CN PC + CG+H C CH
Sbjct: 234 HPGPCPSCPKLVKVRCFCG--CLEDVRRCGFKEFSCNKPCSKFLDCGVHRCIELCHPGAC 407
Query: 710 PPC 712
PPC
Sbjct: 408 PPC 416
Score = 65.5 bits (158), Expect = 1e-10
Identities = 35/90 (38%), Positives = 41/90 (44%), Gaps = 3/90 (3%)
Frame = +3
Query: 575 PHFCSMLCGKKLR--CGQHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGTMP 632
PH C +CG+ L+ CG H C LCH G CP C + + C C + CG
Sbjct: 162 PHSCGEVCGRPLKHSCGHH-CLLLCHPGPCPSCPKLVKVRCFCGC----LEDVRRCGFKE 326
Query: 633 PLCQLPCSVPQPCG-HSGSHSCHFGDCPPC 661
C PCS CG H CH G CPPC
Sbjct: 327 FSCNKPCSKFLDCGVHRCIELCHPGACPPC 416
Score = 50.4 bits (119), Expect = 4e-06
Identities = 41/125 (32%), Positives = 53/125 (41%), Gaps = 12/125 (9%)
Frame = +3
Query: 663 VPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIVKGL 722
+P+S + C S K + P +T C CG+ +PP D P I L
Sbjct: 15 LPISPATASASALWNCPKCRSEYPKSHIP--KTYFC---FCGK-IENPPND--PWI---L 161
Query: 723 RAACGQTCGAP-RSGCRHMCMALCHPGCPCPDARCEFPVTITCSCG-----------RIS 770
+CG+ CG P + C H C+ LCHPG PCP C V + C CG S
Sbjct: 162 PHSCGEVCGRPLKHSCGHHCLLLCHPG-PCPS--CPKLVKVRCFCGCLEDVRRCGFKEFS 332
Query: 771 ANVPC 775
N PC
Sbjct: 333 CNKPC 347
Score = 47.4 bits (111), Expect = 4e-05
Identities = 31/86 (36%), Positives = 35/86 (40%), Gaps = 18/86 (20%)
Frame = +3
Query: 688 CNNPCGRT--RQCGLHACGRSCHSPPCDILPGIVKGLRAACG-----QTCGAPRSGCR-- 738
C CGR CG H C CH PC P +VK +R CG + CG C
Sbjct: 171 CGEVCGRPLKHSCG-HHCLLLCHPGPCPSCPKLVK-VRCFCGCLEDVRRCGFKEFSCNKP 344
Query: 739 ---------HMCMALCHPGCPCPDAR 755
H C+ LCHPG CP R
Sbjct: 345 CSKFLDCGVHRCIELCHPGA-CPPCR 419
>TC215070 similar to UP|Q40336 (Q40336) Proline-rich cell wall protein,
partial (82%)
Length = 1597
Score = 37.0 bits (84), Expect(2) = 0.013
Identities = 50/214 (23%), Positives = 63/214 (29%), Gaps = 36/214 (16%)
Frame = +1
Query: 443 CHPGSCGECEFLP---SRVKACCCGKTKLEDERKSCVDPIPTCSK---------VCSKTL 490
C P C + LP S + C KT + S P C K +CSKT
Sbjct: 226 CTPSLCSKTSSLP*TTSSSTSTPCPKT---SSKASSASPSTLCPKTTSCASYTTICSKTT 396
Query: 491 RCGVHACKET---------------CHVGECPPCKVLISQKCRC----GSTSRTVECYKT 531
C C +T C CP S C S T C KT
Sbjct: 397 NCEPSLCPKTACCASYTTLYP*TTSCDTTICP*TTSGASYTTICP*TTSGASYTTLCAKT 576
Query: 532 TENQKFTC-QKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQ 590
T + C C NC C++ TT + S C K C
Sbjct: 577 THCETTNCLSTSCAKTTNCYTTLCAK------------TTHCETTNCLSTSCAKTTNCYT 720
Query: 591 HVC--ETLCHSGHC--PPCLETIFTDLACACGMT 620
+C T C + +C C T + C T
Sbjct: 721 TLCAKTTHCETTNCLSTSCANTTYCVSTCCTNST 822
Score = 20.8 bits (42), Expect(2) = 0.013
Identities = 8/19 (42%), Positives = 9/19 (47%)
Frame = +2
Query: 623 PPPLPCGTMPPLCQLPCSV 641
PPP P PP Q C +
Sbjct: 881 PPPPPVVPYPPPAQPTCPI 937
>BM270367 similar to PIR|S78452|S784 POU-domain protein rdc-1 - human,
partial (8%)
Length = 405
Score = 37.7 bits (86), Expect = 0.030
Identities = 21/49 (42%), Positives = 23/49 (46%)
Frame = +2
Query: 316 PHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRC 364
PH P P PP F+PPR CPCG RCS R+ G RC
Sbjct: 203 PHPPAAPPAPSPSPPSPTFSPPR-CPCG------RCSPRRGS---GSRC 319
>TC208407 similar to UP|Q84KA9 (Q84KA9) RING/C3HC4/PHD zinc finger-like
protein, partial (36%)
Length = 644
Score = 37.0 bits (84), Expect = 0.051
Identities = 15/46 (32%), Positives = 24/46 (51%)
Frame = +2
Query: 195 KLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTS 240
KL KGT+EC +C + + + C +FH CI +W + T+
Sbjct: 287 KLGKGTLECAVCLNEFEDTETLRLIPKCDHVFHPECIDEWLASHTT 424
>TC210397
Length = 718
Score = 37.0 bits (84), Expect = 0.051
Identities = 13/44 (29%), Positives = 23/44 (51%)
Frame = +2
Query: 191 EIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKW 234
EI+E+L + C +C + + +CY +FH CI++W
Sbjct: 251 EIKERLPQTEETCAVCLSQLSVEDEVRELMNCYHVFHRECIERW 382
>TC206525 weakly similar to UP|Q41042 (Q41042) Pisum sativum L. (clone
na-481-5), partial (20%)
Length = 752
Score = 37.0 bits (84), Expect = 0.051
Identities = 14/43 (32%), Positives = 26/43 (59%), Gaps = 1/43 (2%)
Frame = -1
Query: 90 HRSNHHVAHRVEREHVAHRTDRDHVAHRVE-RERVAHRVEREH 131
H+ +HH H+++R H H+ DR H H+++ R H+++ H
Sbjct: 437 HQIDHHHFHQIDRHHHFHQIDRHHP*HQIDHRHHP*HQIDHHH 309
Score = 32.7 bits (73), Expect = 0.96
Identities = 12/41 (29%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Frame = -1
Query: 125 HRVEREHVAHRMEREHVAHRVEREHVAHRVE-REHVAHRVE 164
H+++ H H+++R H H+++R H H+++ R H H+++
Sbjct: 437 HQIDHHHF-HQIDRHHHFHQIDRHHP*HQIDHRHHP*HQID 318
Score = 30.4 bits (67), Expect = 4.8
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 2/61 (3%)
Frame = -2
Query: 28 KGAGASSSASTTSTTTTASTTTA--AAATTTVDQPTSLHSHEKNTNSDGGSSNQGAVVAP 85
K S + STT+ +T ++TTT+ + ATTT + T+ K+T + S+ P
Sbjct: 481 KVGSISPTPSTTTPSTKSTTTTSTKSTATTTSTKSTATTPSTKSTTATTPSTKSTTTTTP 302
Query: 86 T 86
T
Sbjct: 301 T 299
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 36.6 bits (83), Expect = 0.066
Identities = 26/79 (32%), Positives = 32/79 (39%), Gaps = 6/79 (7%)
Frame = -2
Query: 89 RHRSNHHVAHRVEREHVAH-----RTDRDHVAHRVERERVA-HRVEREHVAHRMEREHVA 142
R R +H R R H+ H RT H H V H +H HR+ H
Sbjct: 576 RRRLHHQRTSRHRRLHICHHHHLLRTCPHHRLHCNP*PDVPIHHSCSKHRLHRLFHHHRN 397
Query: 143 HRVEREHVAHRVEREHVAH 161
HR+ R H HR R H +H
Sbjct: 396 HRLHRSHHHHRSHRLHRSH 340
Score = 32.3 bits (72), Expect = 1.3
Identities = 24/61 (39%), Positives = 31/61 (50%), Gaps = 6/61 (9%)
Frame = -3
Query: 30 AGASSSASTTSTTTTAST----TTAAAATTTVDQPTSLHS--HEKNTNSDGGSSNQGAVV 83
A +S+AS+T+T TTAST TT A A+T TS + GS + GAV
Sbjct: 437 ASTASTASSTTTVTTASTVATTTTVATASTVATSTTSSSPAIASATITASSGSLDAGAVG 258
Query: 84 A 84
A
Sbjct: 257 A 255
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 30.0 bits (66), Expect(2) = 0.076
Identities = 25/89 (28%), Positives = 35/89 (39%), Gaps = 8/89 (8%)
Frame = +2
Query: 89 RHRSNHHVAHRVEREHVAHRTDRDHVAHRVER------ERVAHRVER--EHVAHRMEREH 140
+ +++HH R +R HV + HR ER R+ R E +H HR R
Sbjct: 197 QRQNHHHRLRRRQRRHVGPSLEAPLHLHRQERPLPPRPPRLLRRQEAPPDHQRHRPRR-- 370
Query: 141 VAHRVEREHVAHRVEREHVAHRVERGRGR 169
H V R+ R HR R + R
Sbjct: 371 --HNVPSHRHRPRIRRLRQHHRPPRRKSR 451
Score = 25.0 bits (53), Expect(2) = 0.076
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = +1
Query: 34 SSASTTSTTTTASTTTAAAATTTVDQPTS 62
S TTS ++ ST ++ +TTT PTS
Sbjct: 100 SPTRTTSPASSQSTPSSPPSTTTSPSPTS 186
>TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, complete
Length = 1145
Score = 35.4 bits (80), Expect = 0.15
Identities = 30/126 (23%), Positives = 43/126 (33%)
Frame = +3
Query: 37 STTSTTTTASTTTAAAATTTVDQPTSLHSHEKNTNSDGGSSNQGAVVAPTFARHRSNHHV 96
S T TT T + TTT KNT G +++ + + + +HH
Sbjct: 66 SKTRTTVTRNHKETRTQTTTT----------KNT----GHNHKP*LTTSSLSFSSLHHHH 203
Query: 97 AHRVEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHRVEREHVAHRVER 156
H + HR + H H R H + HR H H + R H + R
Sbjct: 204 HHHESNHGLCHRHNHHHHHHHSHRHNHRHGY---GIHHRHRHRHYDHHIHRHHDCILLLR 374
Query: 157 EHVAHR 162
HR
Sbjct: 375 RRRRHR 392
>TC231630 weakly similar to UP|Q7PR65 (Q7PR65) ENSANGP00000016899 (Fragment),
partial (13%)
Length = 709
Score = 35.4 bits (80), Expect = 0.15
Identities = 16/25 (64%), Positives = 22/25 (88%)
Frame = +1
Query: 32 ASSSASTTSTTTTASTTTAAAATTT 56
A+++A+TT+TTTTA+TTTA TTT
Sbjct: 400 ATTTAATTATTTTANTTTAMGRTTT 474
>TC206524 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 464
Score = 35.4 bits (80), Expect = 0.15
Identities = 16/52 (30%), Positives = 27/52 (51%)
Frame = -3
Query: 93 NHHVAHRVEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHR 144
+HH H+++R H+ H+ D H H H+++R H HR H+ H+
Sbjct: 141 HHHHFHQIDRHHLFHQIDHHHHHHSY------HQIDRHHSFHR----HLYHQ 16
Score = 32.0 bits (71), Expect = 1.6
Identities = 17/50 (34%), Positives = 29/50 (58%)
Frame = -1
Query: 34 SSASTTSTTTTASTTTAAAATTTVDQPTSLHSHEKNTNSDGGSSNQGAVV 83
S+ +TTST +TA+T++ + TTT PT+ T S G S+ + ++
Sbjct: 146 STTTTTSTKSTATTSSTKSTTTTTTTPTT--KSTATTPSTGTSTTRTLII 3
Score = 26.9 bits (58), Expect(2) = 0.22
Identities = 15/47 (31%), Positives = 28/47 (58%), Gaps = 2/47 (4%)
Frame = -1
Query: 28 KGAGASSSASTTSTTTTASTTTAAAATTTV--DQPTSLHSHEKNTNS 72
K S++ ST +++T ++TTT+ +TTT + T+ S K+T +
Sbjct: 221 KVGSISTTPSTATSSTKSATTTSTKSTTTTTSTKSTATTSSTKSTTT 81
Score = 26.6 bits (57), Expect(2) = 0.22
Identities = 10/28 (35%), Positives = 16/28 (56%)
Frame = -3
Query: 90 HRSNHHVAHRVEREHVAHRTDRDHVAHR 117
H +HH H+++R H HR H+ H+
Sbjct: 87 HHHHHHSYHQIDRHHSFHR----HLYHQ 16
>TC209977 UP|Q9HF30 (Q9HF30) Pheromone MFalpha1D (Fragment), partial (41%)
Length = 1421
Score = 34.7 bits (78), Expect = 0.25
Identities = 21/64 (32%), Positives = 26/64 (39%), Gaps = 2/64 (3%)
Frame = -1
Query: 608 TIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGDCPP--CSVPV 665
T+ C S+ PPL +PPLC LP P P SH P CS+P+
Sbjct: 1343 TVHASSFVICSPPSLLPPLLSFFLPPLCDLPLFPPSPSMTVHSHLSDLQVSPSLFCSLPL 1164
Query: 666 SKEC 669
C
Sbjct: 1163 LLVC 1152
>TC232709 weakly similar to UP|Q9LZJ6 (Q9LZJ6) RING-H2 zinc finger protein
ATL5 (At3g62690), partial (19%)
Length = 987
Score = 34.7 bits (78), Expect = 0.25
Identities = 20/81 (24%), Positives = 37/81 (44%), Gaps = 2/81 (2%)
Frame = +3
Query: 183 SSLPQLVQEIQEKLTKG--TVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTS 240
+SLP+L+ + ++ +G V C +C + +C IFH++C+ KW + T+
Sbjct: 318 ASLPKLLYKQTDQFKQGEEVV*CSVCLGTIVEDTISRVLPNCKHIFHVDCVDKWFNSNTT 497
Query: 241 VDLSAEKNLGFNWRCPGCQSV 261
CP C++V
Sbjct: 498 --------------CPICRTV 518
>BE059711
Length = 432
Score = 34.3 bits (77), Expect = 0.33
Identities = 13/70 (18%), Positives = 32/70 (45%)
Frame = +3
Query: 93 NHHVAHRVEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHRVEREHVAH 152
+HH+ H + + H+T H+ ++ + + H+ H+ H + + H+ H+ H
Sbjct: 36 DHHLHH*AKNRDLHHQTRNYHLHYQAKNRDLHHQARDHHLHH*AKNRDLHHQARDHHLHH 215
Query: 153 RVEREHVAHR 162
+ + H+
Sbjct: 216 QAPGRLLHHQ 245
Score = 31.2 bits (69), Expect = 2.8
Identities = 17/70 (24%), Positives = 24/70 (34%)
Frame = +3
Query: 98 HRVEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHRVEREHVAHRVERE 157
H R+H H ++ H R H + H R+H H + H R+
Sbjct: 21 HYQARDHHLHH*AKNRDLHHQTRNYHLHYQAKNRDLHHQARDHHLHH*AKNRDLHHQARD 200
Query: 158 HVAHRVERGR 167
H H GR
Sbjct: 201 HHLHHQAPGR 230
>TC217193 similar to UP|Q7XJN2 (Q7XJN2) At2g40650 protein, partial (88%)
Length = 1691
Score = 34.3 bits (77), Expect = 0.33
Identities = 27/99 (27%), Positives = 40/99 (40%)
Frame = +3
Query: 84 APTFARHRSNHHVAHRVEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAH 143
+PT R R H +HR DRD+ +RER R +R+
Sbjct: 861 SPTRERDRDRRHDSHRHRDRDYDRDYDRDY-----DRERGRGRDRDRDRERDRDRDRYRL 1025
Query: 144 RVEREHVAHRVEREHVAHRVERGRGRSGNMAGRQYGSRD 182
R E+++ R RE +R RGR + + + SRD
Sbjct: 1026REEKDYGRDREGRERERREKDRDRGRRRSHSRSRSRSRD 1142
>TC229486 similar to UP|Q76KV2 (Q76KV2) DNA binding with one finger 2
protein, partial (21%)
Length = 1297
Score = 34.3 bits (77), Expect = 0.33
Identities = 16/27 (59%), Positives = 20/27 (73%)
Frame = +1
Query: 30 AGASSSASTTSTTTTASTTTAAAATTT 56
+ +S + TSTTTTA+TT A AATTT
Sbjct: 115 SSTTSDVTLTSTTTTAATTAATAATTT 195
>TC205952
Length = 1414
Score = 34.3 bits (77), Expect = 0.33
Identities = 20/44 (45%), Positives = 25/44 (56%)
Frame = +3
Query: 18 HRPPRQEWIPKGAGASSSASTTSTTTTASTTTAAAATTTVDQPT 61
H P I A AS S + TT +AS TTAAAA + VD+P+
Sbjct: 903 HPSPASCPIGSNATASKSIWSLPTTISASATTAAAAASYVDEPS 1034
>TC230110 similar to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP4-like,
partial (16%)
Length = 998
Score = 33.9 bits (76), Expect = 0.43
Identities = 18/64 (28%), Positives = 33/64 (51%), Gaps = 2/64 (3%)
Frame = +3
Query: 33 SSSASTTSTTTTASTTTAAAATTT--VDQPTSLHSHEKNTNSDGGSSNQGAVVAPTFARH 90
+S +S+ S++ TA+ TAA TTT V P +L+ N N++ + + ++ P +
Sbjct: 114 NSHSSSGSSSLTAAAATAATTTTTEAVSAPETLNLDSNNNNNNNNNMQESKLLIPALETN 293
Query: 91 RSNH 94
H
Sbjct: 294 PLEH 305
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.134 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 77,196,018
Number of Sequences: 63676
Number of extensions: 1705278
Number of successful extensions: 12739
Number of sequences better than 10.0: 159
Number of HSP's better than 10.0 without gapping: 10676
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12283
length of query: 1173
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1065
effective length of database: 5,762,624
effective search space: 6137194560
effective search space used: 6137194560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC141436.6


