

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141436.1 + phase: 0
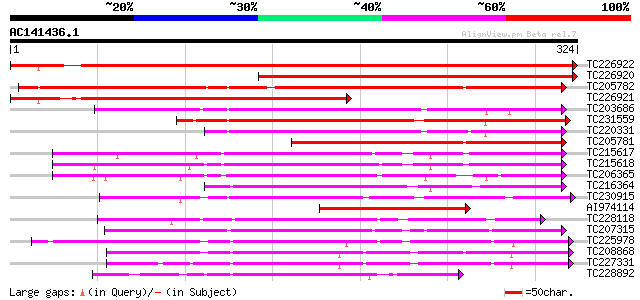
(324 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%) 491 e-139
TC226920 weakly similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial... 330 5e-91
TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, pa... 268 3e-72
TC226921 similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein p... 242 2e-64
TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete 196 9e-51
TC231559 homologue to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (71%) 171 4e-43
TC220331 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partia... 161 4e-40
TC205781 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, pa... 160 6e-40
TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 140 6e-34
TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 140 1e-33
TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, parti... 129 2e-30
TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%) 122 3e-28
TC230915 weakly similar to PIR|T10690|T10690 serine/threonine-sp... 109 1e-24
AI974114 108 3e-24
TC228118 similar to UP|KP19_ARATH (Q39030) Serine/threonine-prot... 105 2e-23
TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kina... 102 3e-22
TC225978 similar to UP|Q9M6N2 (Q9M6N2) Abscisic acid-activated p... 100 7e-22
TC208868 UP|Q39868 (Q39868) Protein kinase , complete 97 8e-21
TC227331 similar to UP|Q43465 (Q43465) Protein kinase 2, partial... 97 8e-21
TC228892 UP|Q9XF25 (Q9XF25) SNF-1-like serine/threonine protein ... 97 1e-20
>TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%)
Length = 1190
Score = 491 bits (1265), Expect = e-139
Identities = 239/326 (73%), Positives = 270/326 (82%), Gaps = 2/326 (0%)
Frame = +1
Query: 1 MALVHRRNSPKLRLP--EISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEK 58
MALV R P LRLP E S+ R RFP+PLP TTA A GD I+A D EK
Sbjct: 73 MALVRHRRHPNLRLPLPEPSERRLRFPLPLPPT---------TTAKPASGDAIAAADLEK 225
Query: 59 LSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKY 118
L++LGHGNGGTVYKVRHK TS YALKI H D+D T RRRAL+E +ILRR TDC +VV++
Sbjct: 226 LAILGHGNGGTVYKVRHKATSATYALKIIHSDTDATRRRRALSETSILRRVTDCPHVVRF 405
Query: 119 HGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHR 178
H SFEKP+GDV ILMEYMD G+LETAL +GTFSE +L+ VARD+L GL YLHARNIAHR
Sbjct: 406 HSSFEKPSGDVAILMEYMDGGTLETALAASGTFSEERLAKVARDVLEGLAYLHARNIAHR 585
Query: 179 DIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGF 238
DIKP+NILVN + +VKIADFGVSK M R+LEACNSYVGTCAYMSP+RFDPE YGGNYNGF
Sbjct: 586 DIKPANILVNSEGDVKIADFGVSKLMCRSLEACNSYVGTCAYMSPDRFDPEAYGGNYNGF 765
Query: 239 SADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECC 298
+ADIWSLGLTLFELYVG+FPFLQ+GQRPDWA+LMCAICF DPPSLPETAS EFR+FVECC
Sbjct: 766 AADIWSLGLTLFELYVGHFPFLQAGQRPDWATLMCAICFGDPPSLPETASPEFRDFVECC 945
Query: 299 LKKESGERWSAAQLLTHPFLCKDMES 324
LKKESGERW+ AQLLTHPF+C D ++
Sbjct: 946 LKKESGERWTTAQLLTHPFVCNDPDT 1023
>TC226920 weakly similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (49%)
Length = 722
Score = 330 bits (846), Expect = 5e-91
Identities = 154/182 (84%), Positives = 168/182 (91%)
Frame = +2
Query: 143 TALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSK 202
TAL T GTFSE +L+ VARD+L GL YLHARNIAHRDIKP+NILVN + EVKIADFGVSK
Sbjct: 20 TALATGGTFSEERLAKVARDVLEGLAYLHARNIAHRDIKPANILVNSEGEVKIADFGVSK 199
Query: 203 FMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQS 262
M RTLEACNSYVGTCAYMSP+RFDPE YGGNYNGF+ADIWSLGLTLFELYVG+FPFLQ+
Sbjct: 200 LMCRTLEACNSYVGTCAYMSPDRFDPEAYGGNYNGFAADIWSLGLTLFELYVGHFPFLQA 379
Query: 263 GQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFLCKDM 322
GQRPDWA+LMCAICFSDPPSLPETAS EF +FVECCLKKESGERW+AAQLLTHPF+CKD
Sbjct: 380 GQRPDWATLMCAICFSDPPSLPETASPEFHDFVECCLKKESGERWTAAQLLTHPFVCKDP 559
Query: 323 ES 324
E+
Sbjct: 560 ET 565
>TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (91%)
Length = 1377
Score = 268 bits (684), Expect = 3e-72
Identities = 137/313 (43%), Positives = 200/313 (63%)
Frame = +2
Query: 6 RRNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHG 65
RR L LP+ D P+PLP + + S + A I + E+L+ +G G
Sbjct: 134 RRKDLTLPLPQ-RDTNLAVPLPLPPSTAPAAAASGGASQQAAQQVIPFSELERLNRIGSG 310
Query: 66 NGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKP 125
+GGTVYKV H+ + +YALK+ + + + RR+ E+ ILR D NVVK H +++
Sbjct: 311 SGGTVYKVVHRTSGRVYALKVIYGHHEESVRRQIHREIQILRDVDDA-NVVKCHEMYDQ- 484
Query: 126 TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNI 185
++ +L+E+MD GSLE T E +L+ ++R IL GL YLH R+I HRDIKPSN+
Sbjct: 485 NSEIQVLLEFMDGGSLEGKHIT----QEQQLADLSRQILRGLAYLHRRHIVHRDIKPSNL 652
Query: 186 LVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSL 245
L+N + +VKIADFGV + + +T++ CNS VGT AYMSPER + ++ G Y+ ++ DIWS
Sbjct: 653 LINSRKQVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDINDGQYDAYAGDIWSF 832
Query: 246 GLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGE 305
G+++ E Y+G FPF G++ DWASLMCAIC S PP P +AS F++F+ CL+++
Sbjct: 833 GVSILEFYMGRFPF-AVGRQGDWASLMCAICMSQPPEAPPSASPHFKDFILRCLQRDPSR 1009
Query: 306 RWSAAQLLTHPFL 318
RWSA++LL HPF+
Sbjct: 1010RWSASRLLEHPFI 1048
>TC226921 similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein precursor,
partial (5%)
Length = 641
Score = 242 bits (617), Expect = 2e-64
Identities = 126/197 (63%), Positives = 148/197 (74%), Gaps = 2/197 (1%)
Frame = +1
Query: 1 MALVHRRNSPKLRLP--EISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEK 58
MALV R P LRLP E S+ RPRFP+PLP P+T T A + GD I++ D EK
Sbjct: 37 MALVRHRRHPNLRLPLPEPSERRPRFPLPLP------PTT--TIAKPSAGDTIASADLEK 192
Query: 59 LSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKY 118
L+VLGHGNGGTVYKVRHK TS YALKI H D+D TTRRRA +E +ILRRATDC +VV++
Sbjct: 193 LAVLGHGNGGTVYKVRHKTTSATYALKIIHSDADATTRRRAFSETSILRRATDCPHVVRF 372
Query: 119 HGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHR 178
HGSFE P+GDV ILMEYMD G+LETAL T GTFSE +L+ VARD+L GLTYLHARNIAHR
Sbjct: 373 HGSFENPSGDVAILMEYMDGGTLETALATGGTFSEERLAKVARDVLKGLTYLHARNIAHR 552
Query: 179 DIKPSNILVNIKNEVKI 195
D++P K EV++
Sbjct: 553 DVEPRAY**TTKEEVRV 603
>TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete
Length = 1502
Score = 196 bits (499), Expect = 9e-51
Identities = 111/275 (40%), Positives = 161/275 (58%), Gaps = 5/275 (1%)
Frame = +2
Query: 49 DNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRR 108
+ ++ D + + V+G GNGG V V+HK TS +ALK+ + + + R++ E+ I ++
Sbjct: 275 NQLTLADIDVIKVVGKGNGGVVQLVQHKWTSQFFALKVIQMNIEESMRKQIAQELKINQQ 454
Query: 109 ATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLT 168
A C VV + SF + G + I++EYMD GSL LK T E L+ + + +L GL
Sbjct: 455 A-QCPYVVVCYQSFYE-NGVISIILEYMDGGSLADLLKKVKTIPEDYLAAICKQVLKGLV 628
Query: 169 YLH-ARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFD 227
YLH ++I HRD+KPSN+L+N EVKI DFGVS M T N+++GT YMSPER +
Sbjct: 629 YLHHEKHIIHRDLKPSNLLINHIGEVKITDFGVSAIMESTSGQANTFIGTYNYMSPERIN 808
Query: 228 PEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASL--MCAICFSDPPSLP- 284
G NY +DIWSLGL L E +G FP+ Q W S+ + PP +P
Sbjct: 809 GSQRGYNY---KSDIWSLGLILLECALGRFPYAPPDQSETWESIFELIETIVDKPPPIPP 979
Query: 285 -ETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
E S+EF +F+ CL+K+ +R SA +L+ HPF+
Sbjct: 980 SEQFSTEFCSFISACLQKDPKDRLSAQELMAHPFV 1084
>TC231559 homologue to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (71%)
Length = 867
Score = 171 bits (433), Expect = 4e-43
Identities = 97/230 (42%), Positives = 141/230 (61%), Gaps = 5/230 (2%)
Frame = +2
Query: 96 RRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESK 155
R++ + E+ I +A+ C +VV + SF G + +++EYMD GSL +K T E
Sbjct: 32 RKQIVQELKI-NQASQCPHVVVCYHSFYH-NGVISLVLEYMDRGSLADVIKQVKTILEPY 205
Query: 156 LSTVARDILNGLTYLH-ARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSY 214
L+ V + +L GL YLH R++ HRDIKPSN+LVN K EVKI DFGVS + ++ +++
Sbjct: 206 LAVVFKQVLQGLVYLHNERHVIHRDIKPSNLLVNHKGEVKITDFGVSAMLASSMGQRDTF 385
Query: 215 VGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWAS---L 271
VGT YMSPER Y +S+DIWSLG+ + E +G FP++QS + W S L
Sbjct: 386 VGTYNYMSPERISGSTY-----DYSSDIWSLGMVVLECAIGRFPYIQSEDQQSWPSFYEL 550
Query: 272 MCAICFSDPPSL-PETASSEFRNFVECCLKKESGERWSAAQLLTHPFLCK 320
+ AI S PPS P+ S EF +FV C++K+ +R ++ +LL HPF+ K
Sbjct: 551 LAAIVESPPPSAPPDQFSPEFCSFVSSCIQKDPRDRLTSLKLLDHPFIKK 700
>TC220331 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial (62%)
Length = 669
Score = 161 bits (407), Expect = 4e-40
Identities = 94/212 (44%), Positives = 124/212 (58%), Gaps = 5/212 (2%)
Frame = +1
Query: 112 CTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLH 171
C VV + SF G + I++EYMD GSLE L T ES LS + + +L GL YLH
Sbjct: 4 CPYVVVCYNSFYH-NGVISIILEYMDGGSLEDLLSKVKTIPESYLSAICKQVLKGLMYLH 180
Query: 172 -ARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEV 230
A++I HRD+KPSN+L+N + EVKI DFGVS M T N+++GT +YMSPER
Sbjct: 181 YAKHIIHRDLKPSNLLINHRGEVKITDFGVSVIMENTSGQANTFIGTYSYMSPERIIGNQ 360
Query: 231 YGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWAS---LMCAICFSDPPSLP-ET 286
+G NY +DIWSLGL L + G FP+ R W + L+ I PS P +
Sbjct: 361 HGYNY---KSDIWSLGLILLKCATGQFPYTPP-DREGWENIFQLIEVIVEKPSPSAPSDD 528
Query: 287 ASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
S EF +F+ CL+K G+R SA L+ HPF+
Sbjct: 529 FSPEFCSFISACLQKNPGDRPSARDLINHPFI 624
>TC205781 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (47%)
Length = 935
Score = 160 bits (406), Expect = 6e-40
Identities = 73/157 (46%), Positives = 102/157 (64%)
Frame = +2
Query: 162 DILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYM 221
D L G H I HRDIKP N + + KIA FGV + + +T++ CN VGT A M
Sbjct: 8 DFLGGWGNWHRGXIVHRDIKPXNXXXXSRKQGKIAXFGVGRILNQTMDPCNXSVGTIANM 187
Query: 222 SPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPP 281
SPER + ++ G Y+ ++ DIWS G+++ E Y+G FPF G++ DWASLMCAIC S PP
Sbjct: 188 SPERINTDINDGQYDAYAGDIWSFGVSILEFYMGRFPF-AVGRQGDWASLMCAICMSQPP 364
Query: 282 SLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
P +AS F++F+ CL+++ RWSA++LL HPF+
Sbjct: 365 EAPPSASPHFKDFILRCLQRDPSRRWSASRLLEHPFI 475
>TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (51%)
Length = 1451
Score = 140 bits (354), Expect = 6e-34
Identities = 95/302 (31%), Positives = 155/302 (50%), Gaps = 8/302 (2%)
Frame = +3
Query: 25 PVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLS---VLGHGNGGTVYKVRHKLTSII 81
P+PLP PS+S + G + G+ K +LG G G VY + + +
Sbjct: 552 PLPLPPGSPTSPSSSLPSTRANGMTEHTTGNLSKWKKGKLLGRGTFGHVYLGFNSDSGQL 731
Query: 82 YALKINHYDSDPTTRRRALTEVNI---LRRATDCTNVVKYHGSFEKPTGDVCILMEYMDS 138
A+K D + + L ++N L N+V+Y+GS + + + +EY+
Sbjct: 732 CAIKEVRVVCDDQSSKECLKQLNQEIHLLSQLSHPNIVQYYGS-DLGEETLSVYLEYVSG 908
Query: 139 GSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADF 198
GS+ L+ G F E + R I++GL+YLH RN HRDIK +NILV+ E+K+ADF
Sbjct: 909 GSIHKLLQEYGAFKEPVIQNYTRQIVSGLSYLHGRNTVHRDIKGANILVDPNGEIKLADF 1088
Query: 199 GVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFS--ADIWSLGLTLFELYVGY 256
G++K + + + S+ G+ +M+PE N NG+S DIWSLG T+ E+
Sbjct: 1089GMAKHINSS-SSMLSFKGSPYWMAPE------VVMNTNGYSLPVDIWSLGCTILEMATSK 1247
Query: 257 FPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHP 316
P+ Q A++ D P +P+ SS+ +NF++ CL+++ R +A +L+ HP
Sbjct: 1248PPW---NQYEGAAAIFKIGNSRDMPEIPDHLSSDAKNFIQLCLQRDPSARPTAQKLIEHP 1418
Query: 317 FL 318
F+
Sbjct: 1419FI 1424
>TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (73%)
Length = 2291
Score = 140 bits (352), Expect = 1e-33
Identities = 98/303 (32%), Positives = 153/303 (50%), Gaps = 9/303 (2%)
Frame = +1
Query: 25 PVPLPANIYKQPSTSATTASVAG---GDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSII 81
P+PLP PS+ + A G + + K +LG G G VY + +
Sbjct: 514 PLPLPPGSPTSPSSVLSNARANGHLENATSNVSKWRKGKLLGRGTFGHVYLGFNSENGQM 693
Query: 82 YALKINHYDSDPTTRRRALT----EVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMD 137
A+K D T + L E+N+L + + N+V+YHGS E + + +EY+
Sbjct: 694 CAIKEVKVVFDDHTSKECLKQLNQEINLLNQLSH-PNIVQYHGS-ELVEESLSVYLEYVS 867
Query: 138 SGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIAD 197
GS+ L+ G F E + R I++GL YLH RN HRDIK +NILV+ E+K+AD
Sbjct: 868 GGSIHKLLQEYGPFKEPVIQNYTRQIVSGLAYLHGRNTVHRDIKGANILVDPNGEIKLAD 1047
Query: 198 FGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFS--ADIWSLGLTLFELYVG 255
FG++K + + S+ G+ +M+PE N NG+S DIWSLG T+ E+
Sbjct: 1048FGMAKHINSSASML-SFKGSPYWMAPE------VVMNTNGYSLPVDIWSLGCTIIEMATS 1206
Query: 256 YFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTH 315
P+ Q A++ D P +PE S++ + F++ CL+++ R +A +LL H
Sbjct: 1207KPPW---NQYEGVAAIFKIGNSKDMPEIPEHLSNDAKKFIKLCLQRDPLARPTAQKLLDH 1377
Query: 316 PFL 318
PF+
Sbjct: 1378PFI 1386
>TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, partial (37%)
Length = 1866
Score = 129 bits (324), Expect = 2e-30
Identities = 103/314 (32%), Positives = 162/314 (50%), Gaps = 20/314 (6%)
Frame = +1
Query: 25 PVPLPANIYKQPSTSATTASVA----GGDNISA--GDFEKLSVLGHGNGGTVYKVRHKLT 78
P+PLP Q + S+ + +N+ + G ++K ++G G+ G+VY + T
Sbjct: 325 PLPLPPKASPQTAHSSPQHQPSIVHLNTENLPSMKGQWQKGKLIGRGSYGSVYHATNLET 504
Query: 79 SIIYALK-INHYDSDPTTR---RRALTEVNILRRATDCTNVVKYHGSFEKPTGD-VCILM 133
ALK ++ + DP + ++ E+ ILR+ N+V+Y+GS + GD + I M
Sbjct: 505 GASCALKEVDLFPDDPKSADCIKQLEQEIRILRQLHH-PNIVQYYGS--EIVGDRLYIYM 675
Query: 134 EYMDSGSLETAL-KTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNE 192
EY+ GSL + + G +ES + R IL+GL YLH HRDIK +N+LV+
Sbjct: 676 EYVHPGSLHKFMHEHCGAMTESVVRNFTRHILSGLAYLHGTKTIHRDIKGANLLVDASGS 855
Query: 193 VKIADFGVSKFM-GRTLEACNSYVGTCAYMSPERFDPEVYGGNYN--GFSADIWSLGLTL 249
VK+ADFGVSK + ++ E S G+ +M+PE + + + DIWSLG T+
Sbjct: 856 VKLADFGVSKILTEKSYEL--SLKGSPYWMAPELMKAAIKKESSPDIAMAIDIWSLGCTI 1029
Query: 250 FELYVGYFPFLQSGQRPDWASL-----MCAICFSDPPSLPETASSEFRNFVECCLKKESG 304
E+ G +P W+ M + P LPE+ SSE ++F++ C ++
Sbjct: 1030IEMLTG---------KPPWSEFEGPQAMFKVLHKS-PDLPESLSSEGQDFLQQCFRRNPA 1179
Query: 305 ERWSAAQLLTHPFL 318
ER SAA LLTH F+
Sbjct: 1180ERPSAAVLLTHAFV 1221
>TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%)
Length = 2224
Score = 122 bits (305), Expect = 3e-28
Identities = 74/209 (35%), Positives = 114/209 (54%), Gaps = 2/209 (0%)
Frame = +1
Query: 112 CTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLH 171
C + +Y+GS+ T + I+MEYM GS+ +++ E ++ + RD+L+ + YLH
Sbjct: 25 CPYITEYYGSYLNQT-KLWIIMEYMAGGSVADLIQSGPPLDEMSIACILRDLLHAVDYLH 201
Query: 172 ARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVY 231
+ HRDIK +NIL++ +VK+ADFGVS + RT+ ++VGT +M+PE
Sbjct: 202 SEGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQ---- 369
Query: 232 GGNYNGFS--ADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASS 289
N +G++ ADIWSLG+T E+ G P ++ I +PP L + S
Sbjct: 370 --NTDGYNEKADIWSLGITAIEMAKGEPPLADLHP----MRVLFIIPRENPPQLDDHFSR 531
Query: 290 EFRNFVECCLKKESGERWSAAQLLTHPFL 318
+ FV CLKK ER SA +LL F+
Sbjct: 532 PLKEFVSLCLKKVPAERPSAKELLKDRFI 618
>TC230915 weakly similar to PIR|T10690|T10690 serine/threonine-specific
protein kinase homolog T16I18.40 - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (90%)
Length = 1174
Score = 109 bits (273), Expect = 1e-24
Identities = 79/278 (28%), Positives = 131/278 (46%), Gaps = 6/278 (2%)
Frame = +3
Query: 52 SAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTR-----RRALTEVNIL 106
S DFE LG G G VY R + + ALK+ + R RR + L
Sbjct: 66 SLEDFEIGKPLGRGKFGRVYVAREVKSKFVVALKVIFKEQIDKYRVHHQLRREMEIQTSL 245
Query: 107 RRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNG 166
R A N+++ +G F V +++EY G L L+ G +E + +T +
Sbjct: 246 RHA----NILRLYGGFHD-ADRVFLILEYAHKGELNKELRKKGHLTEKQAATYIVSLTKA 410
Query: 167 LTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERF 226
L Y H +++ HRDIKP N+L + + +KIADFG S ++ ++ GT Y++PE
Sbjct: 411 LAYCHEKHVIHRDIKPENLLPDHEGRLKIADFGWSV---QSRSKRHTMCGTLDYLAPEMA 581
Query: 227 DPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLM-CAICFSDPPSLPE 285
+ + + ++ D W+LG+ +E G PF Q + +M + F PS+
Sbjct: 582 ENKAH-----DYAVDNWTLGIRCYESLYGAPPFEAESQSDTFKRIMKVDLSFPSTPSV-- 740
Query: 286 TASSEFRNFVECCLKKESGERWSAAQLLTHPFLCKDME 323
S E +N + L K+S R S +++ HP++ K+ +
Sbjct: 741 --SIEAKNLISRLLVKDSSRRLSLQKIMEHPWIIKNAD 848
>AI974114
Length = 259
Score = 108 bits (270), Expect = 3e-24
Identities = 56/86 (65%), Positives = 64/86 (74%)
Frame = -1
Query: 178 RDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNG 237
RDI PS V + +VKI DF VSK M R+L AC SYVGTCAYMSP+RFD E Y GNYNG
Sbjct: 259 RDI*PSIS*VRSEVDVKITDFAVSKLMCRSL*ACTSYVGTCAYMSPDRFDSEAYDGNYNG 80
Query: 238 FSADIWSLGLTLFELYVGYFPFLQSG 263
F+A I S+GLTL LY G+FPFL +G
Sbjct: 79 FAAYICSVGLTLSVLYGGHFPFLLAG 2
>TC228118 similar to UP|KP19_ARATH (Q39030) Serine/threonine-protein kinase
AtPK19 (Ribosomal-protein S6 kinase homolog) , partial
(65%)
Length = 1305
Score = 105 bits (263), Expect = 2e-23
Identities = 80/258 (31%), Positives = 126/258 (48%), Gaps = 2/258 (0%)
Frame = +1
Query: 51 ISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDS--DPTTRRRALTEVNILRR 108
+S DFE L V+G G VY+VR K TS IYA+K+ D + E +I +
Sbjct: 151 VSIDDFEILKVVGQGAFAKVYQVRKKDTSEIYAMKVMRKDKIMEKNHAEYMKAERDIWTK 330
Query: 109 ATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLT 168
+ VV+ SF+ + +++++++ G L L G F E +I++ ++
Sbjct: 331 I-EHPFVVQLRYSFQTKYR-LYLVLDFVNGGHLFFQLYHQGLFREDLARIYTAEIVSAVS 504
Query: 169 YLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDP 228
+LHA I HRD+KP NIL++ V + DFG++K + + NS GT YM+ P
Sbjct: 505 HLHANGIMHRDLKPENILLDADGHVMLTDFGLAKQFEESTRS-NSMCGTLEYMA-----P 666
Query: 229 EVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETAS 288
E+ G + +AD WS+G+ LFE G PF + ++ D LP S
Sbjct: 667 EIILGKGHDKAADWWSVGVLLFETLTGKPPFCGGNRDKIQQKIV-----KDKIKLPAFLS 831
Query: 289 SEFRNFVECCLKKESGER 306
SE + ++ L+KE R
Sbjct: 832 SEAHSLLKGLLQKEQARR 885
>TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1063
Score = 102 bits (253), Expect = 3e-22
Identities = 70/266 (26%), Positives = 134/266 (50%), Gaps = 2/266 (0%)
Frame = +3
Query: 55 DFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALT-EVNILRRATDCT 113
+++ L LG G GTV++ H+ ++ YA K+ RR + E + +
Sbjct: 54 EYQVLEELGRGRFGTVFRCFHRTSNKFYAAKLIEKRRLLNEDRRCIEMEAKAMSFLSPHP 233
Query: 114 NVVKYHGSFEKPTGDVC-ILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHA 172
N+++ +FE D C I++E +L + G +E +++ + +L + + HA
Sbjct: 234 NILQIMDAFED--ADSCSIVLELCQPHTLLDRIAAQGPLTEPHAASLLKQLLEAVAHCHA 407
Query: 173 RNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYG 232
+ +AHRDIKP NIL + N++K++DFG ++++G + + VGT Y++PE + G
Sbjct: 408 QGLAHRDIKPENILFDEGNKLKLSDFGSAEWLGEG-SSMSGVVGTPYYVAPE----VIMG 572
Query: 233 GNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFR 292
Y+ D+WS G+ L+ + G+ PF + S++ A P + + S+ +
Sbjct: 573 REYDE-KVDVWSSGVILYAMLAGFPPFYGESAPEIFESVLRA-NLRFPSLIFSSVSAPAK 746
Query: 293 NFVECCLKKESGERWSAAQLLTHPFL 318
+ + + ++ R SA Q L HP++
Sbjct: 747 DLLRKMISRDPSNRISAHQALRHPWI 824
>TC225978 similar to UP|Q9M6N2 (Q9M6N2) Abscisic acid-activated protein
kinase, partial (98%)
Length = 1606
Score = 100 bits (250), Expect = 7e-22
Identities = 87/315 (27%), Positives = 143/315 (44%), Gaps = 5/315 (1%)
Frame = +2
Query: 13 RLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGNGGTVYK 72
R P +S H P P P I + AT V + ++ + +G GN G
Sbjct: 101 RSPNLSPHPQ--PPPYPPLILQPMDRPATGPGVDMPIMHDSDRYDFVRDIGSGNFGVARL 274
Query: 73 VRHKLTSIIYALK-INHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCI 131
+R K T + A+K I D +R + LR N++++ PT + I
Sbjct: 275 MRDKQTQELVAVKYIERGDKIDENVKREIINHRSLRHP----NIIRFKEVILTPT-HLAI 439
Query: 132 LMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKN 191
+MEY G L + G F+E + + +++G++Y HA + HRD+K N L++
Sbjct: 440 VMEYASGGELFEKICNAGHFNEDEARFFFQQLISGVSYCHAMEVCHRDLKLENTLLDGSP 619
Query: 192 --EVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTL 249
+KI DFG SK + S VGT AY++PE + Y+G AD+WS G+TL
Sbjct: 620 ALHLKICDFGYSKSSVLHSQP-KSTVGTPAYIAPE----VLLKQEYDGKIADVWSCGVTL 784
Query: 250 FELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPET--ASSEFRNFVECCLKKESGERW 307
F + VG +PF D+ + + S S+P+ S E R+ + + ER
Sbjct: 785 FVMLVGSYPFEDPNDPKDFRKTIQRV-LSVQYSIPDNVQVSPECRHLISRIFVFDPAERI 961
Query: 308 SAAQLLTHPFLCKDM 322
+ ++L + + K++
Sbjct: 962 TIPEILQNEWFLKNL 1006
>TC208868 UP|Q39868 (Q39868) Protein kinase , complete
Length = 1217
Score = 97.4 bits (241), Expect = 8e-21
Identities = 77/272 (28%), Positives = 125/272 (45%), Gaps = 5/272 (1%)
Frame = +1
Query: 56 FEKLSVLGHGNGGTVYKVRHKLTSIIYALK-INHYDSDPTTRRRALTEVNILRRATDCTN 114
+E L LG GN G + K T + A+K I +R + LR N
Sbjct: 103 YETLKELGSGNFGVARLAKDKETGELVAIKYIERGKKIDANVQREIVNHRSLRHP----N 270
Query: 115 VVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARN 174
++++ F PT + I++EY G L + G SE + + +++G++Y H+
Sbjct: 271 IIRFKEVFLTPT-HLAIVLEYAAGGELFERICNAGRLSEDEARCFFQQLISGVSYCHSMQ 447
Query: 175 IAHRDIKPSNILV--NIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYG 232
I HRD+K N L+ N +KI DFG SK S VGT AY++PE +
Sbjct: 448 ICHRDLKLENTLLDGNPAPRLKICDFGFSK-SALLHSQPKSTVGTPAYIAPEVLSRK--- 615
Query: 233 GNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPE--TASSE 290
Y+G AD+WS G+TL+ + VG +PF + + P S ++P+ S E
Sbjct: 616 -EYDGKVADVWSCGVTLYVMLVGAYPF-EDPEDPKNFRKSIGRIMSVQYAIPDYVRVSKE 789
Query: 291 FRNFVECCLKKESGERWSAAQLLTHPFLCKDM 322
R+ + C +R S +++ H + K++
Sbjct: 790 CRHLISCIFVANPAKRISISEIKQHLWFRKNL 885
>TC227331 similar to UP|Q43465 (Q43465) Protein kinase 2, partial (88%)
Length = 1334
Score = 97.4 bits (241), Expect = 8e-21
Identities = 76/271 (28%), Positives = 128/271 (47%), Gaps = 4/271 (1%)
Frame = +2
Query: 56 FEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNV 115
+E L LG GN G + K T + A+K + + E+ I R+ N+
Sbjct: 71 YEPLKELGAGNFGVARLAKDKKTGELVAVK--YIERGKKIDENVQREI-INHRSLRHPNI 241
Query: 116 VKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNI 175
+++ PT + I++EY G L + T G FSE + + +++G++Y H+ I
Sbjct: 242 IRFKEVLLTPT-HLAIVLEYASGGELFERICTAGRFSEDEARYFFQQLISGVSYCHSMEI 418
Query: 176 AHRDIKPSNILV--NIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGG 233
HRD+K N L+ N +KI DFG SK S VGT AY++PE +
Sbjct: 419 CHRDLKLENTLLDGNPSPRLKICDFGYSK-SALLHSQPKSTVGTPAYIAPEVLSRK---- 583
Query: 234 NYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPE--TASSEF 291
Y+G +D+WS G+TL+ + VG +PF ++ + I S+P+ SS+
Sbjct: 584 EYDGKISDVWSCGVTLYVMLVGAYPFEDPEDPRNFRKTIGRI-IGIQYSIPDYVRVSSDC 760
Query: 292 RNFVECCLKKESGERWSAAQLLTHPFLCKDM 322
RN + + +R + ++ +P+ K+M
Sbjct: 761 RNLLSRIFVADPAKRITIPEIKQYPWFLKNM 853
>TC228892 UP|Q9XF25 (Q9XF25) SNF-1-like serine/threonine protein kinase,
complete
Length = 1931
Score = 96.7 bits (239), Expect = 1e-20
Identities = 69/216 (31%), Positives = 105/216 (47%), Gaps = 4/216 (1%)
Frame = +1
Query: 48 GDNISAGDFEKLSVLGHGNGGTVYKVRHKLT-SIIYALKINHYDSDPTTRRRALTEVNIL 106
G + G F K+ + H G HK+ I+ KI + + + RR E+ IL
Sbjct: 145 GKTLGIGSFGKVKIAEHVRTG------HKVAIKILNRHKIKNMEMEEKVRR----EIKIL 294
Query: 107 RRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNG 166
R ++++ + E PT D+ ++MEY+ SG L + G E + + I++G
Sbjct: 295 RLFMH-HHIIRLYEVVETPT-DIYVVMEYVKSGELFDYIVEKGRLQEDEARHFFQQIISG 468
Query: 167 LTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFM--GRTLEACNSYVGTCAYMSPE 224
+ Y H + HRD+KP N+L++ K +KIADFG+S M G L+ +C SP
Sbjct: 469 VEYCHRNMVVHRDLKPENLLLDSKFNIKIADFGLSNIMRDGHFLKT------SCG--SPN 624
Query: 225 RFDPEVYGGN-YNGFSADIWSLGLTLFELYVGYFPF 259
PEV G Y G D+WS G+ L+ L G PF
Sbjct: 625 YAAPEVISGKLYAGPEVDVWSCGVILYALLCGTLPF 732
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,241,065
Number of Sequences: 63676
Number of extensions: 223095
Number of successful extensions: 2103
Number of sequences better than 10.0: 670
Number of HSP's better than 10.0 without gapping: 1859
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1893
length of query: 324
length of database: 12,639,632
effective HSP length: 97
effective length of query: 227
effective length of database: 6,463,060
effective search space: 1467114620
effective search space used: 1467114620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC141436.1


