

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141414.11 - phase: 0
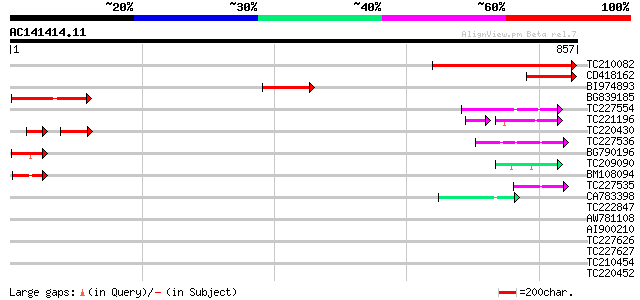
(857 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC210082 396 e-110
CD418162 145 7e-35
BI974893 100 2e-21
BG839185 89 1e-17
TC227554 similar to UP|Q84RR2 (Q84RR2) ARIADNE-like protein ARI2... 67 4e-11
TC221196 weakly similar to UP|Q9FHR4 (Q9FHR4) Mutator-like trans... 61 3e-10
TC220430 similar to UP|Q9FF84 (Q9FF84) ATP-dependent RNA helicas... 44 4e-10
TC227536 similar to UP|Q84RR0 (Q84RR0) ARIADNE-like protein ARI7... 60 4e-09
BG790196 57 5e-08
TC209090 56 8e-08
BM108094 55 1e-07
TC227535 similar to UP|Q84RR0 (Q84RR0) ARIADNE-like protein ARI7... 47 4e-05
CA783398 44 2e-04
TC222847 similar to UP|Q6YUI7 (Q6YUI7) RNA helicase-like, partia... 39 0.010
AW781108 38 0.022
AI900210 similar to PIR|T48023|T480 ATP-dependent RNA helicase-l... 33 0.41
TC227626 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (34%) 33 0.41
TC227627 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (49%) 33 0.41
TC210454 weakly similar to GB|AAP68217.1|31711722|BT008778 At3g0... 33 0.54
TC220452 33 0.70
>TC210082
Length = 779
Score = 396 bits (1018), Expect = e-110
Identities = 168/217 (77%), Positives = 196/217 (89%)
Frame = +1
Query: 640 ERFDTGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNH 699
ERF GPSCPICLCEVEDGY+LEGCGHLFC+ C+VEQ ESAIKNQG+FP+ C H+ CG+
Sbjct: 19 ERFGNGPSCPICLCEVEDGYRLEGCGHLFCRMCLVEQFESAIKNQGTFPVCCTHRDCGDP 198
Query: 700 ILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVC 759
ILL D R+LL DKLE+LFRASLGAFVA+S GTYRFCPSPDCPSIYRVADP++A PFVC
Sbjct: 199 ILLTDLRSLLFGDKLEDLFRASLGAFVATSGGTYRFCPSPDCPSIYRVADPESAGEPFVC 378
Query: 760 GACYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDG 819
ACYSETCTRCH+EYHPY+SCERY++FK+DPDSSL++WC+GKEQVK C ACG+VIEKVDG
Sbjct: 379 RACYSETCTRCHLEYHPYLSCERYKEFKEDPDSSLKEWCRGKEQVKCCSACGYVIEKVDG 558
Query: 820 CNHIECKCGKHICWVCLEFFTTSGECYSHMDTIHLGV 856
CNH+ECKCGKH+CWVCLEFF+TS +CY+H+ TIHL +
Sbjct: 559 CNHVECKCGKHVCWVCLEFFSTSNDCYNHVRTIHLTI 669
>CD418162
Length = 364
Score = 145 bits (366), Expect = 7e-35
Identities = 56/75 (74%), Positives = 70/75 (92%)
Frame = -1
Query: 782 RYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIECKCGKHICWVCLEFFTT 841
RY++FK+DPDSSL++WC+GKEQVK C ACG+VIEKVDGCNH+ECKCGKH+CWVCLEFF+T
Sbjct: 355 RYQEFKEDPDSSLKEWCRGKEQVKCCSACGYVIEKVDGCNHVECKCGKHVCWVCLEFFST 176
Query: 842 SGECYSHMDTIHLGV 856
S +CY+H+ TIHL +
Sbjct: 175 SNDCYNHLRTIHLAI 131
>BI974893
Length = 245
Score = 100 bits (250), Expect = 2e-21
Identities = 52/78 (66%), Positives = 61/78 (77%)
Frame = +2
Query: 383 MNALIHFDGRLHLEAAKALEKIDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQL 442
+ ALI FDGRLH +AAKALE+I+GKVLPG SWQKIK Q+LFHS+L F PVY VI QL
Sbjct: 8 LRALITFDGRLH*QAAKALEQIEGKVLPGCLSWQKIKCQQLFHSSLTFPTPVYRVITEQL 187
Query: 443 EKVLARFNNLEGLEWKLD 460
++VLA F N +GLE LD
Sbjct: 188 DEVLASFLNPKGLECNLD 241
>BG839185
Length = 744
Score = 88.6 bits (218), Expect = 1e-17
Identities = 49/123 (39%), Positives = 76/123 (60%), Gaps = 3/123 (2%)
Frame = +3
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESG---GCY 60
Q+ ++ GETGSGK+TQ+ Q+L ++G + + CTQPRR+AA S+A RV +E G G
Sbjct: 243 QVLIIDGETGSGKTTQIPQYLHEAGYTKHGMVACTQPRRLAAISVAARVSKEMGVKLGHE 422
Query: 61 EDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLL 120
SI+ F + I +MTD LL+ ++ + + S +IVDEAHER+++TD+L
Sbjct: 423 VGYSIR----FEDCTTDKTVIKYMTDGMLLREFLGEPDLASYSVLIVDEAHERTLSTDIL 590
Query: 121 LAL 123
+
Sbjct: 591 FGI 599
>TC227554 similar to UP|Q84RR2 (Q84RR2) ARIADNE-like protein ARI2, partial
(61%)
Length = 1678
Score = 66.6 bits (161), Expect = 4e-11
Identities = 49/160 (30%), Positives = 70/160 (43%), Gaps = 7/160 (4%)
Frame = +1
Query: 683 NQG-SFPIRCAHQGCGNHILLVDFRTLLSNDK---LEELFRASLGAFVASSSGTYRFCPS 738
N+G S IRC C + RTLLS + E+ R L +++ + ++CPS
Sbjct: 19 NEGQSKRIRCMEHKCNSICDEAVVRTLLSREHSHMAEKYERFLLESYIEDNKRV-KWCPS 195
Query: 739 -PDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLR 795
P C + RV D + CG + C C E H SC + + K +S
Sbjct: 196 TPHCGNAIRVEDDELCEVECSCGVQF---CFSCLSEAHSPCSCLMWELWAKKCRDESETV 366
Query: 796 DWCKGKEQVKNCPACGHVIEKVDGCNHIECKCGKHICWVC 835
+W K CP C +EK GCN + C CG+ CW+C
Sbjct: 367 NWIT--VHTKPCPKCHKPVEKNGGCNLVSCICGQAFCWLC 480
Score = 34.7 bits (78), Expect = 0.18
Identities = 22/82 (26%), Positives = 35/82 (41%), Gaps = 4/82 (4%)
Frame = +1
Query: 759 CGACYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPA---CGHVIE 815
C + E R + E+Y +F L + + ++VK CP+ CG+ I
Sbjct: 61 CNSICDEAVVRTLLSREHSHMAEKYERFL------LESYIEDNKRVKWCPSTPHCGNAIR 222
Query: 816 -KVDGCNHIECKCGKHICWVCL 836
+ D +EC CG C+ CL
Sbjct: 223 VEDDELCEVECSCGVQFCFSCL 288
>TC221196 weakly similar to UP|Q9FHR4 (Q9FHR4) Mutator-like transposase,
partial (20%)
Length = 758
Score = 61.2 bits (147), Expect(2) = 3e-10
Identities = 29/114 (25%), Positives = 49/114 (42%), Gaps = 13/114 (11%)
Frame = +1
Query: 735 FCPSPDCPSIYR----------VADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYR 784
+CP P C ++ + S P +C C C C + +H ++C Y+
Sbjct: 136 YCPYPTCSALMSKTEVLEYSKDITGQSEQSEPKICLKCRGLFCFNCKVPWHSGMTCNTYK 315
Query: 785 QFKDDP---DSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIECKCGKHICWVC 835
+ P D L+ + + + C C H+IE +GC H+ C+CG C+ C
Sbjct: 316 RMNPIPPAEDLKLK-FLASRSLWQQCLKCNHMIELAEGCYHMTCRCGYEFCYNC 474
Score = 22.7 bits (47), Expect(2) = 3e-10
Identities = 10/37 (27%), Positives = 18/37 (48%)
Frame = +3
Query: 690 RCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFV 726
+C HQGC +L+ + L+ E + + L A +
Sbjct: 6 KCPHQGCKYELLVDSCQKFLTQKLTETMQQRKLEASI 116
>TC220430 similar to UP|Q9FF84 (Q9FF84) ATP-dependent RNA helicase A-like
protein, partial (24%)
Length = 915
Score = 43.5 bits (101), Expect(2) = 4e-10
Identities = 19/47 (40%), Positives = 31/47 (65%)
Frame = +1
Query: 78 DSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
++ ++F T LL+ +SD+N GI+ + VDE HER +N D LL ++
Sbjct: 178 NTHLLFCTSGILLRRLLSDRNPNGITHVFVDEIHERGMNEDFLLIVL 318
Score = 39.7 bits (91), Expect(2) = 4e-10
Identities = 19/32 (59%), Positives = 24/32 (74%)
Frame = +2
Query: 26 DSGVGANESIVCTQPRRIAAKSLAERVREESG 57
+SG GA SI+CTQPRRI+ ++AERV E G
Sbjct: 11 ESGRGAFCSIICTQPRRISVMAVAERVSAERG 106
>TC227536 similar to UP|Q84RR0 (Q84RR0) ARIADNE-like protein ARI7, partial
(52%)
Length = 893
Score = 60.1 bits (144), Expect = 4e-09
Identities = 45/147 (30%), Positives = 68/147 (45%), Gaps = 6/147 (4%)
Frame = +3
Query: 704 DFRTLLSNDKLEELF-RASLGAFVASSSGTYRFCPSPDCPSIYRVA-DPDTASAPFVCGA 761
D LL++D+ +E + R L +++ + T ++CP+P C Y V D + + C
Sbjct: 15 DMINLLASDEDKEKYDRYLLRSYIEDNKKT-KWCPAPGCE--YAVTFDAGSGNYDVSCLC 185
Query: 762 CYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDG 819
YS C C E H V C ++ K+ +S +W K CP C IEK G
Sbjct: 186 SYS-FCWNCTEEAHRPVDCGTVSKWILKNSAESENMNWILANS--KPCPKCKRPIEKNQG 356
Query: 820 CNHIEC--KCGKHICWVCLEFFTTSGE 844
C H+ C C CW+CL ++ GE
Sbjct: 357 CMHMTCTPPCKFEFCWLCLGAWSDHGE 437
>BG790196
Length = 304
Score = 56.6 bits (135), Expect = 5e-08
Identities = 28/58 (48%), Positives = 41/58 (70%), Gaps = 4/58 (6%)
Frame = +1
Query: 4 QITVLIGETGSGKSTQLVQFLADSGV----GANESIVCTQPRRIAAKSLAERVREESG 57
Q+ V+ GETG GK+TQL QF+ + + GA+ +I+CTQPRR++A S+A R+ E G
Sbjct: 85 QVLVVSGETGCGKTTQLPQFILEEEISCLRGADCNIICTQPRRVSAISVAARISAERG 258
>TC209090
Length = 1011
Score = 55.8 bits (133), Expect = 8e-08
Identities = 31/114 (27%), Positives = 45/114 (39%), Gaps = 13/114 (11%)
Frame = +1
Query: 735 FCPSPDCPSIYRVADPDTASAP---------FVCGACYSETCTRCHIEYHPYVSCERYRQ 785
+CP P+C + + +A A C C C C + +H +SC Y+
Sbjct: 55 YCPFPNCSVLLDPHECSSARASSSSQSDNSCIECPVCRRFICVDCKVPWHSSMSCLEYQN 234
Query: 786 F----KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIECKCGKHICWVC 835
+D D +L + K K C C IE GC H+ C CG C+ C
Sbjct: 235 LPEKERDVSDITLHRLAQNKRW-KRCQQCRRTIELTQGCYHMTCWCGHEFCYSC 393
>BM108094
Length = 635
Score = 55.1 bits (131), Expect = 1e-07
Identities = 31/53 (58%), Positives = 35/53 (65%)
Frame = +1
Query: 5 ITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESG 57
+ V+IGETGSGKSTQL Q L G G I TQPRR+AA S+A RV E G
Sbjct: 46 VVVVIGETGSGKSTQLSQMLHRRGYG---KIAITQPRRVAAVSVARRVAHELG 195
>TC227535 similar to UP|Q84RR0 (Q84RR0) ARIADNE-like protein ARI7, partial
(54%)
Length = 1486
Score = 47.0 bits (110), Expect = 4e-05
Identities = 27/87 (31%), Positives = 37/87 (42%), Gaps = 4/87 (4%)
Frame = +2
Query: 762 CYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDG 819
C C C E H V C ++ K+ +S +W K CP C IEK G
Sbjct: 38 CSYGFCWNCTEEAHRPVDCGTVAKWILKNSAESENMNWILANS--KPCPKCKRPIEKNQG 211
Query: 820 CNHIEC--KCGKHICWVCLEFFTTSGE 844
C H+ C C CW+C+ ++ GE
Sbjct: 212 CMHMTCTPPCKFEFCWLCVGAWSDHGE 292
>CA783398
Length = 457
Score = 44.3 bits (103), Expect = 2e-04
Identities = 30/125 (24%), Positives = 49/125 (39%), Gaps = 2/125 (1%)
Frame = +3
Query: 648 CPICL--CEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHILLVDF 705
C IC D + + GC H++C SC+ + ES ++ + I C GC + D
Sbjct: 90 CEICTETKTARDSFSIIGCHHVYCNSCVAQYVESKLE-ENIVSIPCPVPGCRGLLEADDC 266
Query: 706 RTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGACYSE 765
R +L+ + + A +A+ Y CP DC + + C C
Sbjct: 267 REILAPRVFDRWGKVLCEAVIAAEEKFY--CPFADCSVMLIRGIEENNIREAECPNCRRL 440
Query: 766 TCTRC 770
C +C
Sbjct: 441 FCAQC 455
>TC222847 similar to UP|Q6YUI7 (Q6YUI7) RNA helicase-like, partial (12%)
Length = 477
Score = 38.9 bits (89), Expect = 0.010
Identities = 16/34 (47%), Positives = 24/34 (70%)
Frame = +2
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVC 37
Q+ V++GETGSGK+TQ+ Q+L ++G I C
Sbjct: 374 QVLVIVGETGSGKTTQIPQYLHEAGYTKRGMIAC 475
>AW781108
Length = 438
Score = 37.7 bits (86), Expect = 0.022
Identities = 18/36 (50%), Positives = 26/36 (72%)
Frame = +1
Query: 89 LLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
LL+ M D++ +GI+ IIVDE HERS+ D LL ++
Sbjct: 10 LLRKLMGDQSLSGITHIIVDEVHERSLLGDFLLIVL 117
>AI900210 similar to PIR|T48023|T480 ATP-dependent RNA helicase-like protein
- Arabidopsis thaliana, partial (10%)
Length = 411
Score = 33.5 bits (75), Expect = 0.41
Identities = 13/24 (54%), Positives = 20/24 (83%)
Frame = +1
Query: 4 QITVLIGETGSGKSTQLVQFLADS 27
Q +L+GETGSGK+TQ+ QF+ ++
Sbjct: 319 QTLILVGETGSGKTTQIPQFVLEA 390
>TC227626 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (34%)
Length = 834
Score = 33.5 bits (75), Expect = 0.41
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 2/30 (6%)
Frame = +2
Query: 648 CPICLCEVEDGYQLEG--CGHLFCQSCMVE 675
C ICLC EDG +L C H F SC+V+
Sbjct: 308 CCICLCSYEDGAELHALPCNHHFHSSCIVK 397
>TC227627 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (49%)
Length = 967
Score = 33.5 bits (75), Expect = 0.41
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 2/30 (6%)
Frame = +3
Query: 648 CPICLCEVEDGYQLEG--CGHLFCQSCMVE 675
C ICLC EDG +L C H F SC+V+
Sbjct: 489 CCICLCSYEDGAELHALPCNHHFHSSCIVK 578
>TC210454 weakly similar to GB|AAP68217.1|31711722|BT008778 At3g05250
{Arabidopsis thaliana;} , partial (25%)
Length = 449
Score = 33.1 bits (74), Expect = 0.54
Identities = 18/69 (26%), Positives = 30/69 (43%), Gaps = 2/69 (2%)
Frame = +1
Query: 625 EEITFEIARLSNPSSERFDTGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQ 684
++ E+ +P E D+ CPICL L+ C H FC +C++ +
Sbjct: 1 DDAAIEVRNHVSPEPEVEDSSCCCPICLGSFLQLSYLDKCFHKFCFNCILRWTKVVASKH 180
Query: 685 GSFP--IRC 691
S P ++C
Sbjct: 181 RSPPSSVKC 207
>TC220452
Length = 537
Score = 32.7 bits (73), Expect = 0.70
Identities = 23/80 (28%), Positives = 34/80 (41%), Gaps = 9/80 (11%)
Frame = +1
Query: 602 DLRLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSSERFDTG---------PSCPICL 652
+++LN R + H L E I + AR++ P ++ P CPICL
Sbjct: 34 NIQLNDRESVLHQH-KRPLPPPNEVIHHDNARMAIPHMPNLESAESTAPAAAEPVCPICL 210
Query: 653 CEVEDGYQLEGCGHLFCQSC 672
+D GCGH C+ C
Sbjct: 211 TNPKD--MAFGCGHTTCKEC 264
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,991,853
Number of Sequences: 63676
Number of extensions: 685391
Number of successful extensions: 3494
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 3425
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3486
length of query: 857
length of database: 12,639,632
effective HSP length: 105
effective length of query: 752
effective length of database: 5,953,652
effective search space: 4477146304
effective search space used: 4477146304
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC141414.11


