

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141323.12 + phase: 0 /pseudo
(133 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
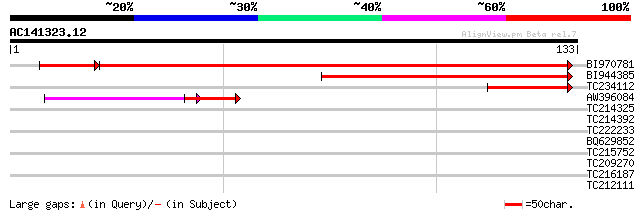
Score E
Sequences producing significant alignments: (bits) Value
BI970781 similar to GP|17381240|gb AT5g09790/F17I14_20 {Arabidop... 179 2e-46
BI944385 similar to GP|17381240|gb| AT5g09790/F17I14_20 {Arabido... 101 9e-23
TC234112 similar to UP|Q8VZJ1 (Q8VZJ1) AT5g09790/F17I14_20, part... 40 4e-04
AW396084 weakly similar to GP|17381240|gb| AT5g09790/F17I14_20 {... 35 5e-04
TC214325 similar to GB|AAP31966.1|30387601|BT006622 At3g19640 {A... 29 0.56
TC214392 similar to GB|AAP31966.1|30387601|BT006622 At3g19640 {A... 29 0.56
TC222233 similar to UP|Q6IMT1 (Q6IMT1) SAB, partial (7%) 28 1.6
BQ629852 27 2.1
TC215752 weakly similar to UP|Q8YQN5 (Q8YQN5) All3785 protein, p... 26 4.8
TC209270 26 4.8
TC216187 26 6.2
TC212111 weakly similar to GB|AAP37775.1|30725506|BT008416 At1g5... 25 8.1
>BI970781 similar to GP|17381240|gb AT5g09790/F17I14_20 {Arabidopsis
thaliana}, partial (39%)
Length = 753
Score = 179 bits (454), Expect(2) = 2e-46
Identities = 86/111 (77%), Positives = 99/111 (88%)
Frame = -2
Query: 22 SMMERGECPLLMVVFDHAEGYTVEADKSIKDLTVIREYVGDIDFLKNREYDDGDRIMTLL 81
S+MERGE LMVVFD EG+TVEAD+SIKDLT+I EYVGD+DFLKNRE DDGD IMTLL
Sbjct: 629 SVMERGEXXPLMVVFDPLEGFTVEADRSIKDLTIITEYVGDVDFLKNRENDDGDSIMTLL 450
Query: 82 SASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNKKQNMKCVRFNIGGECR 132
SAS+PS++LV+ PDKRSNIA FI GI+NHTPEG KKQN+KCVRF++GGECR
Sbjct: 449 SASDPSRTLVICPDKRSNIARFINGINNHTPEGKKKQNLKCVRFDVGGECR 297
Score = 22.3 bits (46), Expect(2) = 2e-46
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = -1
Query: 8 VLSKEDFETLNLSR 21
V SKED ETLNL +
Sbjct: 669 VXSKEDTETLNLCK 628
>BI944385 similar to GP|17381240|gb| AT5g09790/F17I14_20 {Arabidopsis
thaliana}, partial (23%)
Length = 394
Score = 101 bits (252), Expect = 9e-23
Identities = 46/59 (77%), Positives = 54/59 (90%)
Frame = +2
Query: 74 GDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNKKQNMKCVRFNIGGECR 132
GD IMTLLSAS+PS++LV+ PDKRSNIA FI GI+NHTPEG KKQN+KCVRF++GGECR
Sbjct: 143 GDSIMTLLSASDPSRTLVICPDKRSNIARFINGINNHTPEGKKKQNLKCVRFDVGGECR 319
>TC234112 similar to UP|Q8VZJ1 (Q8VZJ1) AT5g09790/F17I14_20, partial (14%)
Length = 581
Score = 39.7 bits (91), Expect = 4e-04
Identities = 15/20 (75%), Positives = 17/20 (85%)
Frame = +2
Query: 113 EGNKKQNMKCVRFNIGGECR 132
EG KKQN KCVR+N+ GECR
Sbjct: 125 EGRKKQNCKCVRYNVNGECR 184
>AW396084 weakly similar to GP|17381240|gb| AT5g09790/F17I14_20 {Arabidopsis
thaliana}, partial (6%)
Length = 433
Score = 34.7 bits (78), Expect(2) = 5e-04
Identities = 18/37 (48%), Positives = 22/37 (58%)
Frame = +2
Query: 9 LSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVE 45
LSKED ETL ++ +RGE P MV H EG +E
Sbjct: 206 LSKEDMETLEQCIALSKRGEFPPFMVFMIHREGTHLE 316
Score = 23.9 bits (50), Expect(2) = 5e-04
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +1
Query: 42 YTVEADKSIKDLT 54
YTVEAD IKD+T
Sbjct: 394 YTVEADDLIKDMT 432
>TC214325 similar to GB|AAP31966.1|30387601|BT006622 At3g19640 {Arabidopsis
thaliana;} , partial (48%)
Length = 1186
Score = 29.3 bits (64), Expect = 0.56
Identities = 16/53 (30%), Positives = 26/53 (48%)
Frame = +1
Query: 40 EGYTVEADKSIKDLTVIREYVGDIDFLKNREYDDGDRIMTLLSASNPSQSLVV 92
E Y V+ D ++ L+ +REYV D + N DD + + + +LVV
Sbjct: 625 EAYFVQIDGTLNKLSTLREYVDDTEDYINIMLDDKQNHLLQMGVMLTTATLVV 783
>TC214392 similar to GB|AAP31966.1|30387601|BT006622 At3g19640 {Arabidopsis
thaliana;} , partial (67%)
Length = 1330
Score = 29.3 bits (64), Expect = 0.56
Identities = 16/53 (30%), Positives = 26/53 (48%)
Frame = +2
Query: 40 EGYTVEADKSIKDLTVIREYVGDIDFLKNREYDDGDRIMTLLSASNPSQSLVV 92
E Y V+ D ++ L+ +REYV D + N DD + + + +LVV
Sbjct: 884 EAYFVQIDGTLNKLSTLREYVDDTEDYINIMLDDKQNHLLQMGVMLTTATLVV 1042
>TC222233 similar to UP|Q6IMT1 (Q6IMT1) SAB, partial (7%)
Length = 597
Score = 27.7 bits (60), Expect = 1.6
Identities = 23/65 (35%), Positives = 34/65 (51%), Gaps = 9/65 (13%)
Frame = -3
Query: 36 FDHAEGYTVEADKSIKDLTVIREYVGDIDFLKNREYDDG------DRIMTLLSASN---P 86
F H++G ADKSI DLT+ R+++ + NR IM+L+SAS P
Sbjct: 370 FPHSDG-GFHADKSISDLTLGRQFLTTKYLVVNRAMPTSL*SRSKSYIMSLISASAKDLP 194
Query: 87 SQSLV 91
S S++
Sbjct: 193 STSIL 179
>BQ629852
Length = 396
Score = 27.3 bits (59), Expect = 2.1
Identities = 9/29 (31%), Positives = 18/29 (62%)
Frame = +1
Query: 28 ECPLLMVVFDHAEGYTVEADKSIKDLTVI 56
+C L+++ FDH Y ++ KSI +++
Sbjct: 289 QCQLILLPFDHCTHYVIQGPKSILHKSIL 375
>TC215752 weakly similar to UP|Q8YQN5 (Q8YQN5) All3785 protein, partial (7%)
Length = 1310
Score = 26.2 bits (56), Expect = 4.8
Identities = 13/35 (37%), Positives = 20/35 (57%)
Frame = +3
Query: 82 SASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNK 116
S+ NPSQS V FP K+ + F G + +G++
Sbjct: 210 SSPNPSQSSVTFPKKKWRLLWFRHGNTSSKTDGSE 314
>TC209270
Length = 660
Score = 26.2 bits (56), Expect = 4.8
Identities = 9/29 (31%), Positives = 18/29 (62%)
Frame = -3
Query: 28 ECPLLMVVFDHAEGYTVEADKSIKDLTVI 56
+C L+++ FDH Y ++ KSI +++
Sbjct: 334 QCRLILLPFDHYTHYVIQGPKSILHKSIL 248
>TC216187
Length = 977
Score = 25.8 bits (55), Expect = 6.2
Identities = 9/20 (45%), Positives = 13/20 (65%)
Frame = -1
Query: 105 TGIDNHTPEGNKKQNMKCVR 124
T + H P GNK++N+ VR
Sbjct: 266 TSAETHRPTGNKRRNVSAVR 207
>TC212111 weakly similar to GB|AAP37775.1|30725506|BT008416 At1g51350
{Arabidopsis thaliana;} , partial (24%)
Length = 518
Score = 25.4 bits (54), Expect = 8.1
Identities = 15/53 (28%), Positives = 26/53 (48%)
Frame = +1
Query: 3 VCISHVLSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVEADKSIKDLTV 55
+C+ V + L+ R M ER CPL+ ++ D + V A +I ++ V
Sbjct: 52 ICLRSV--SRSIKNLSAGRFMNERVVCPLVQLLSDLSTSVQVAALGAISNIVV 204
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,293,956
Number of Sequences: 63676
Number of extensions: 55378
Number of successful extensions: 231
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 230
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 231
length of query: 133
length of database: 12,639,632
effective HSP length: 87
effective length of query: 46
effective length of database: 7,099,820
effective search space: 326591720
effective search space used: 326591720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 53 (25.0 bits)
Medicago: description of AC141323.12


