

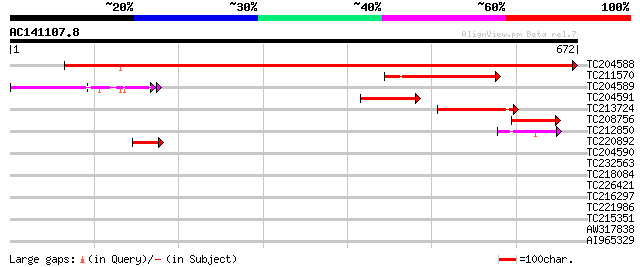
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.8 + phase: 0
(672 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204588 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 1071 0.0
TC211570 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 195 6e-50
TC204589 similar to UP|Q76LU4 (Q76LU4) Alpha-L-arabinofuranosida... 137 1e-32
TC204591 homologue to UP|Q6RIB6 (Q6RIB6) Cytosolic malate dehydr... 126 3e-29
TC213724 weakly similar to UP|Q944F3 (Q944F3) Arabinosidase ARA-... 92 1e-18
TC208756 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 61 2e-09
TC212850 weakly similar to UP|Q76LU3 (Q76LU3) Alpha-L-arabinofur... 56 6e-08
TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-rel... 45 8e-05
TC204590 41 0.002
TC232563 weakly similar to GB|AAQ89617.1|37202004|BT010595 At3g1... 32 0.91
TC218084 32 1.2
TC226421 similar to UP|Q9ZST3 (Q9ZST3) Pyrophosphate-dependent p... 31 2.0
TC216297 similar to UP|Q93X09 (Q93X09) Vacuolar sorting receptor... 30 3.5
TC221986 weakly similar to GB|BAC16401.1|23237826|AP003749 selen... 29 5.9
TC215351 29 7.7
AW317838 similar to SP|P05046|LEC_S Lectin precursor (Agglutinin... 29 7.7
AI965329 29 7.7
>TC204588 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(84%)
Length = 2003
Score = 1071 bits (2770), Expect = 0.0
Identities = 517/611 (84%), Positives = 560/611 (91%), Gaps = 4/611 (0%)
Frame = +2
Query: 66 GAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVALRLEVL 125
G GLWAELVSNRGFEAGGPN PSNIDPWSIIGN + IN+ETDRTSCF+RNKVALRLEVL
Sbjct: 8 GTRGLWAELVSNRGFEAGGPNTPSNIDPWSIIGNESLINIETDRTSCFDRNKVALRLEVL 187
Query: 126 CDGT----CPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSLA 181
CD CP DGVGVYNPGFWGMNIEQGKKYKVVFY RST LNL VSLTGSNGVG+LA
Sbjct: 188 CDSKGDNICPADGVGVYNPGFWGMNIEQGKKYKVVFYVRSTESLNLTVSLTGSNGVGNLA 367
Query: 182 STVITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGF 241
S+VITGSA DFSNWTKVE +L AKAT+ NSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGF
Sbjct: 368 SSVITGSALDFSNWTKVEKILVAKATDHNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGF 547
Query: 242 RSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDDG 301
R+DL++ML LKP FIRFPGGCFVEG++LRNAFRWK ++GPWEERPGHFGDVW YWTDDG
Sbjct: 548 RTDLVEMLTALKPRFIRFPGGCFVEGEWLRNAFRWKTSIGPWEERPGHFGDVWMYWTDDG 727
Query: 302 LGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKWGS 361
LGYYEFLQL+EDLG PIWVFNNGVSHND+VDTSAVLP VQEALDGLEFARGD TSKWGS
Sbjct: 728 LGYYEFLQLAEDLGTAPIWVFNNGVSHNDQVDTSAVLPLVQEALDGLEFARGDRTSKWGS 907
Query: 362 MRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPLD 421
+RAAMGHP PF+L+YVAVGNEDCGKKNYRGNYL+FY AI+ AYPDIQIISNCDGSSRPLD
Sbjct: 908 LRAAMGHPAPFDLRYVAVGNEDCGKKNYRGNYLKFYSAIKSAYPDIQIISNCDGSSRPLD 1087
Query: 422 HPADMYDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAGF 481
HPADMYDYH+YTNANDMFSR+ F+ +R+GPKAFVSEYAVTG DAG GSLLAALAEAGF
Sbjct: 1088HPADMYDYHVYTNANDMFSRANAFDHTSRNGPKAFVSEYAVTGKDAGTGSLLAALAEAGF 1267
Query: 482 LIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATL 541
LIGLEKNSD++ MASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYW+QLFFSES+GATL
Sbjct: 1268LIGLEKNSDVIQMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWVQLFFSESSGATL 1447
Query: 542 LNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSG 601
L++SLQ +SNSLVASAIT+QNSVDKKNYIRIK VNFGT+AVNLKIS +GL+PNSLQ SG
Sbjct: 1448LSTSLQAPSSNSLVASAITFQNSVDKKNYIRIKVVNFGTTAVNLKISLDGLEPNSLQLSG 1627
Query: 602 STKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLKESSNLKM 661
STKTVLTS N+MDENSFSQPKKV+PIQSLLQ+VGKDMNV VPP SFTSFDL+K+SS LKM
Sbjct: 1628STKTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKDMNVTVPPRSFTSFDLVKQSSTLKM 1807
Query: 662 LESDSSSWSSI 672
SDSS+ SSI
Sbjct: 1808AGSDSSTRSSI 1840
>TC211570 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(20%)
Length = 414
Score = 195 bits (495), Expect = 6e-50
Identities = 95/137 (69%), Positives = 114/137 (82%)
Frame = +3
Query: 445 FNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAGFLIGLEKNSDIVHMASYAPLFVNA 504
F+ +RSGPKAFVSEYAV +DA G+LLAA+AEA FLIGLEKNSDI M SYAPLF+N
Sbjct: 6 FDNTSRSGPKAFVSEYAVK-SDAANGNLLAAVAEAAFLIGLEKNSDIGEMVSYAPLFLNI 182
Query: 505 NDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAITWQNS 564
ND+RW PDAIVFNS+Q+YGTPSYW+Q F ES+GAT ++S+L TT+SN L AS I WQ+S
Sbjct: 183 NDKRWIPDAIVFNSYQVYGTPSYWVQKLFIESSGATFIDSTLSTTSSNKLAASVIIWQDS 362
Query: 565 VDKKNYIRIKAVNFGTS 581
DK+NY+RIKAVNFG +
Sbjct: 363 SDKRNYLRIKAVNFGAA 413
>TC204589 similar to UP|Q76LU4 (Q76LU4) Alpha-L-arabinofuranosidase, partial
(14%)
Length = 808
Score = 137 bits (346), Expect = 1e-32
Identities = 93/186 (50%), Positives = 105/186 (56%), Gaps = 13/186 (6%)
Frame = +3
Query: 1 MGFSKDSCCVFMLQLLIVVYLVVQCFDVQVQADLNATLVVDASQASGRRIPETLFGIFFE 60
MG SK C V +L L IVV LV QCF + ADL TL V+AS ASG+ IPETLFGIFFE
Sbjct: 222 MGSSKALCSVSLLHLFIVVCLVFQCFATKKHADLTTTLTVNASDASGKPIPETLFGIFFE 401
Query: 61 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYIN----VETDRTSCFERN 116
EINHAGAGGLWAELVSNRGFEAGGPN PSNI +G +N ++ R
Sbjct: 402 EINHAGAGGLWAELVSNRGFEAGGPNTPSNI----ALGQLLEMNH**ILKLIAPLALTRT 569
Query: 117 KVALRLEVLCDGTC---------PTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNL 167
+V D C P DGV V GMNIEQ ++ FY R L +
Sbjct: 570 RVH------SDWRCM*HQX*HIWPADGV-VAITLVLGMNIEQXEEXXXGFYCRQLDHL-I 725
Query: 168 KVSLTG 173
SLTG
Sbjct: 726 YXSLTG 743
Score = 58.5 bits (140), Expect = 9e-09
Identities = 41/90 (45%), Positives = 48/90 (52%), Gaps = 3/90 (3%)
Frame = +2
Query: 93 PWSIIGNATYINVETDRTSCFERNKVALRLEVLCDGTCPTDG---VGVYNPGFWGMNIEQ 149
PWSIIGN + IN+ETDRTSCF+ K ALRLEV G YNPG
Sbjct: 497 PWSIIGNESLINIETDRTSCFDPXKGALRLEVYVTPXVTHLAR*WGGGYNPGSGHEY*AX 676
Query: 150 GKKYKVVFYARSTGPLNLKVSLTGSNGVGS 179
G + + STG LNL + SNGVG+
Sbjct: 677 GGNXXXLLLS-STGSLNLXF-INWSNGVGN 760
>TC204591 homologue to UP|Q6RIB6 (Q6RIB6) Cytosolic malate dehydrogenase ,
partial (84%)
Length = 1604
Score = 126 bits (317), Expect = 3e-29
Identities = 59/71 (83%), Positives = 65/71 (91%)
Frame = +2
Query: 416 SSRPLDHPADMYDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAA 475
+SRPLDHPADMYDYH+YTNANDMFSR+ F+ +R+GPKAFVSEYAVTG DAG GSLLAA
Sbjct: 1391 ASRPLDHPADMYDYHVYTNANDMFSRANAFDHTSRNGPKAFVSEYAVTGKDAGTGSLLAA 1570
Query: 476 LAEAGFLIGLE 486
LAEAGFLIGLE
Sbjct: 1571 LAEAGFLIGLE 1603
>TC213724 weakly similar to UP|Q944F3 (Q944F3) Arabinosidase ARA-1, partial
(10%)
Length = 595
Score = 91.7 bits (226), Expect = 1e-18
Identities = 51/96 (53%), Positives = 62/96 (64%)
Frame = +3
Query: 508 RWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAITWQNSVDK 567
RW+PDAIVFNS Q+Y TPSY +Q F ESNGAT L S LQT +S+ ASAI W+N DK
Sbjct: 141 RWSPDAIVFNSNQVYETPSYRVQFMFRESNGATFLKSQLQTPDPDSVDASAILWKNLQDK 320
Query: 568 KNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSGST 603
K Y++IK + LKISF L P L + +T
Sbjct: 321 KTYLKIKVIQ*RWHQSVLKISF--LLPKKLNNCHTT 422
>TC208756 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(5%)
Length = 544
Score = 60.8 bits (146), Expect = 2e-09
Identities = 31/59 (52%), Positives = 42/59 (70%)
Frame = +1
Query: 595 NSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLL 653
+++Q GST TVLTSTN+MDENSF +PKKV+P S L+ K + V++P +S S D L
Sbjct: 4 SNVQQYGSTITVLTSTNVMDENSFLEPKKVVPQTSPLEIDDKHLIVLLPSYSVISLDFL 180
>TC212850 weakly similar to UP|Q76LU3 (Q76LU3) Alpha-L-arabinofuranosidase,
partial (3%)
Length = 558
Score = 55.8 bits (133), Expect = 6e-08
Identities = 37/111 (33%), Positives = 53/111 (47%), Gaps = 35/111 (31%)
Frame = +2
Query: 579 GTSAVNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQP----------------- 621
G++ +N KIS NG + S ++ +TKTVLTS N +DENS + P
Sbjct: 11 GSNQINFKISLNGFE--SSNATKATKTVLTSKNALDENSLANPTNVHITSFILFLLL*TN 184
Query: 622 ------------------KKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLK 654
+++P +S LQS ++N I+PP S T FDLLK
Sbjct: 185 NTYNN*LS*HLMFICLFCTQIVPKRSPLQSANNEINDILPPVSLTVFDLLK 337
>TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-related
protein, partial (5%)
Length = 647
Score = 45.4 bits (106), Expect = 8e-05
Identities = 22/37 (59%), Positives = 27/37 (72%)
Frame = +3
Query: 146 NIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSLAS 182
NIE+G+KYKVVFYA++ G +NL VS GS LAS
Sbjct: 129 NIEKGQKYKVVFYAKADGAINLDVSFVGSEKGEKLAS 239
>TC204590
Length = 290
Score = 40.8 bits (94), Expect = 0.002
Identities = 19/23 (82%), Positives = 21/23 (90%)
Frame = +2
Query: 650 FDLLKESSNLKMLESDSSSWSSI 672
FDLLK+SSNLKM SDSS+WSSI
Sbjct: 2 FDLLKQSSNLKMAGSDSSTWSSI 70
>TC232563 weakly similar to GB|AAQ89617.1|37202004|BT010595 At3g10650
{Arabidopsis thaliana;} , partial (4%)
Length = 602
Score = 32.0 bits (71), Expect = 0.91
Identities = 27/99 (27%), Positives = 46/99 (46%), Gaps = 3/99 (3%)
Frame = +3
Query: 522 YGTPSYWMQLFFSESNGATLLNSS-LQTTASNSLVASAITWQNSVDK--KNYIRIKAVNF 578
+G+ S +FF S+ T +NSS Q++A + S + Q S + ++V+F
Sbjct: 162 FGSSSAGSGIFFFSSSAMTTVNSSQSQSSAVGASSGSVLGAQASFTSGFATSTQTQSVSF 341
Query: 579 GTSAVNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENS 617
G+SA + +G P S SS + + S + NS
Sbjct: 342 GSSASSSSFGLSGNTPFSSSSSFPSSSPAASAAFLSGNS 458
>TC218084
Length = 932
Score = 31.6 bits (70), Expect = 1.2
Identities = 27/84 (32%), Positives = 43/84 (51%)
Frame = -2
Query: 585 LKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPP 644
L +S L + +S + + ++T T+L DEN+F I + SLL +G N+ PP
Sbjct: 403 LNLSSLSLSVKPVNASCTEENLITFTSLNDENTF------INVSSLL-CLG---NLSTPP 254
Query: 645 HSFTSFDLLKESSNLKMLESDSSS 668
+ SFD S + L+S +SS
Sbjct: 253 PTLVSFDADASQSWVPSLQS*TSS 182
>TC226421 similar to UP|Q9ZST3 (Q9ZST3) Pyrophosphate-dependent
phosphofructokinase beta subunit , partial (95%)
Length = 2056
Score = 30.8 bits (68), Expect = 2.0
Identities = 19/65 (29%), Positives = 27/65 (41%)
Frame = -3
Query: 163 GPLNLKVSLTGSNGVGSLASTVITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGV 222
GP + + G GS ST + WT + T + T P R+ L T +G
Sbjct: 215 GPWRMVKGCLRTEGRGSAWSTRVF--------WTSL*TEAKRPVTEPEGRVPLGTVVEGA 60
Query: 223 IWLDQ 227
IWL +
Sbjct: 59 IWLQR 45
>TC216297 similar to UP|Q93X09 (Q93X09) Vacuolar sorting receptor, partial
(78%)
Length = 1641
Score = 30.0 bits (66), Expect = 3.5
Identities = 13/40 (32%), Positives = 24/40 (59%)
Frame = +1
Query: 398 DAIRRAYPDIQIISNCDGSSRPLDHPADMYDYHIYTNAND 437
D+I++A D ++++ L HP D +Y ++TN+ND
Sbjct: 55 DSIKQALSDGEMVNINLDWRESLPHPDDRVEYELWTNSND 174
>TC221986 weakly similar to GB|BAC16401.1|23237826|AP003749 selenium-binding
protein-like {Oryza sativa (japonica cultivar-group);} ,
partial (10%)
Length = 853
Score = 29.3 bits (64), Expect = 5.9
Identities = 10/22 (45%), Positives = 18/22 (81%)
Frame = -2
Query: 195 WTKVETVLEAKATNPNSRLQLT 216
W K++T+LE+K NP+++L+ T
Sbjct: 84 WLKLKTILESKIMNPSTQLEAT 19
>TC215351
Length = 566
Score = 28.9 bits (63), Expect = 7.7
Identities = 17/51 (33%), Positives = 28/51 (54%)
Frame = +1
Query: 592 LDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIV 642
LDP+SL S + T+ T +M N+F QP I +LL + ++V++
Sbjct: 199 LDPSSLPSQIALTTIFTQKPVMH*NTFHQPLSHNIITNLLSTKLGPLHVVL 351
>AW317838 similar to SP|P05046|LEC_S Lectin precursor (Agglutinin) (SBA).
[Soybean] {Glycine max}, partial (23%)
Length = 202
Score = 28.9 bits (63), Expect = 7.7
Identities = 13/42 (30%), Positives = 25/42 (58%)
Frame = +2
Query: 546 LQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKI 587
L T+ +NS ++ +W N V K+ + ++ GTS+V L++
Sbjct: 32 LLTSKANSAETASFSWNNGVPKQPNMILQGDAIGTSSVKLQL 157
>AI965329
Length = 407
Score = 28.9 bits (63), Expect = 7.7
Identities = 17/51 (33%), Positives = 28/51 (54%)
Frame = +1
Query: 592 LDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIV 642
LDP+SL S + T+ T +M N+F QP I +LL + ++V++
Sbjct: 19 LDPSSLPSQIALTTIFTQKPVMH*NTFHQPLSHNIITNLLSTKLGPLHVVL 171
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,334,844
Number of Sequences: 63676
Number of extensions: 417531
Number of successful extensions: 1832
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 1822
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1830
length of query: 672
length of database: 12,639,632
effective HSP length: 104
effective length of query: 568
effective length of database: 6,017,328
effective search space: 3417842304
effective search space used: 3417842304
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC141107.8


