

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.11 + phase: 0 /pseudo
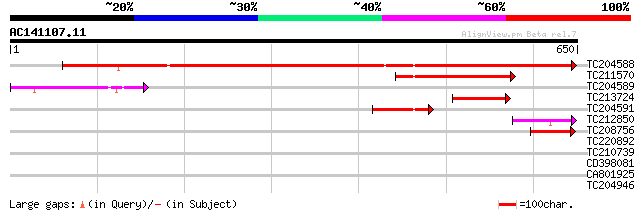
(650 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204588 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 767 0.0
TC211570 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 154 9e-38
TC204589 similar to UP|Q76LU4 (Q76LU4) Alpha-L-arabinofuranosida... 100 3e-21
TC213724 weakly similar to UP|Q944F3 (Q944F3) Arabinosidase ARA-... 97 2e-20
TC204591 homologue to UP|Q6RIB6 (Q6RIB6) Cytosolic malate dehydr... 83 3e-16
TC212850 weakly similar to UP|Q76LU3 (Q76LU3) Alpha-L-arabinofur... 67 2e-11
TC208756 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 58 2e-08
TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-rel... 35 0.14
TC210739 weakly similar to UP|Q8S9K7 (Q8S9K7) At1g48640/F11I4_17... 28 9.8
CD398081 28 9.8
CA801925 28 9.8
TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 28 9.8
>TC204588 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(84%)
Length = 2003
Score = 767 bits (1981), Expect = 0.0
Identities = 374/596 (62%), Positives = 451/596 (74%), Gaps = 7/596 (1%)
Frame = +2
Query: 61 GSGGIWAQLVNNSGFEAAGTRTPSNIFPWTIIGTESSVKLQTELSSCFERNKVALRMDVL 120
G+ G+WA+LV+N GFEA G TPSNI PW+IIG ES + ++T+ +SCF+RNKVALR++VL
Sbjct: 8 GTRGLWAELVSNRGFEAGGPNTPSNIDPWSIIGNESLINIETDRTSCFDRNKVALRLEVL 187
Query: 121 CDK-----CPPDGVGVSNPGFWGMNIVQGKKYKVVFFYRSLGSLDMRVAFRDAISGRILA 175
CD CP DGVGV NPGFWGMNI QGKKYKVVF+ RS SL++ V+ + LA
Sbjct: 188 CDSKGDNICPADGVGVYNPGFWGMNIEQGKKYKVVFYVRSTESLNLTVSLTGSNGVGNLA 367
Query: 176 SSHIIRHKASKKKGSKWQRVQTILEARASSSNSNLTLTTTKEGTVWLDQVSAMPTDTFKG 235
SS I S S W +V+ IL A+A+ NS L LTTT +G +WLDQVSAMP DT+KG
Sbjct: 368 SSVIT---GSALDFSNWTKVEKILVAKATDHNSRLQLTTTTKGVIWLDQVSAMPLDTYKG 538
Query: 236 HGFRKDLVEMLIQLKPAFLRFPGGCFVEGVQLRNAFRWKDSVGPWEQRPGHLNDIWNYWT 295
HGFR DLVEML LKP F+RFPGGCFVEG LRNAFRWK S+GPWE+RPGH D+W YWT
Sbjct: 539 HGFRTDLVEMLTALKPRFIRFPGGCFVEGEWLRNAFRWKTSIGPWEERPGHFGDVWMYWT 718
Query: 296 DDGLGFFEGLQLAEDIGALPIWVFNNGISHSDEVDTSVISPFVKEALEGIEFARGSSTSK 355
DDGLG++E LQLAED+G PIWVFNNG+SH+D+VDTS + P V+EAL+G+EFARG TSK
Sbjct: 719 DDGLGYYEFLQLAEDLGTAPIWVFNNGVSHNDQVDTSAVLPLVQEALDGLEFARGDRTSK 898
Query: 356 WGSVRASMGHPKPFNLKYVAIGNEDCYKKNYYGNYMAFYKAIKKFYPDIQIISNCPAFKT 415
WGS+RA+MGHP PF+L+YVA+GNEDC KKNY GNY+ FY AIK YPDIQIISNC
Sbjct: 899 WGSLRAAMGHPAPFDLRYVAVGNEDCGKKNYRGNYLKFYSAIKSAYPDIQIISNCDGSSR 1078
Query: 416 PLNHPADLYDYHTYPIDARAMFNAYHDFDKSPRNGPKAFVSEYALIGALQAKYGTLLGAV 475
PL+HPAD+YDYH Y +A MF+ + FD + RNGPKAFVSEYA+ G A G+LL A+
Sbjct: 1079PLDHPADMYDYHVY-TNANDMFSRANAFDHTSRNGPKAFVSEYAVTGK-DAGTGSLLAAL 1252
Query: 476 SEAGFLIGLERNSDHVAMASYAPLLVNANDRNWNPDAIVFNSYQAYGTPSYWVTYMFKES 535
+EAGFLIGLE+NSD + MASYAPL VNANDR WNPDAIVFNS+Q YGTPSYWV F ES
Sbjct: 1253AEAGFLIGLEKNSDVIQMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWVQLFFSES 1432
Query: 536 NGATFLNSQLQTPDPGSLIASAILCQSPQNNSTYLKIKIANIGSIPVNLKISLQGYV--S 593
+GAT L++ LQ P SL+ASAI Q+ + Y++IK+ N G+ VNLKISL G S
Sbjct: 1433SGATLLSTSLQAPSSNSLVASAITFQNSVDKKNYIRIKVVNFGTTAVNLKISLDGLEPNS 1612
Query: 594 KNLAGSTKTVLTSGNILDENTFAAPKKIAPQTSPLQNPGNEMNVMIPPVSLTVLDM 649
L+GSTKTVLTS N++DEN+F+ PKK+ P S LQN G +MNV +PP S T D+
Sbjct: 1613LQLSGSTKTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKDMNVTVPPRSFTSFDL 1780
>TC211570 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(20%)
Length = 414
Score = 154 bits (390), Expect = 9e-38
Identities = 79/137 (57%), Positives = 94/137 (67%)
Frame = +3
Query: 443 FDKSPRNGPKAFVSEYALIGALQAKYGTLLGAVSEAGFLIGLERNSDHVAMASYAPLLVN 502
FD + R+GPKAFVSEYA+ A G LL AV+EA FLIGLE+NSD M SYAPL +N
Sbjct: 6 FDNTSRSGPKAFVSEYAVKS--DAANGNLLAAVAEAAFLIGLEKNSDIGEMVSYAPLFLN 179
Query: 503 ANDRNWNPDAIVFNSYQAYGTPSYWVTYMFKESNGATFLNSQLQTPDPGSLIASAILCQS 562
ND+ W PDAIVFNSYQ YGTPSYWV +F ES+GATF++S L T L AS I+ Q
Sbjct: 180 INDKRWIPDAIVFNSYQVYGTPSYWVQKLFIESSGATFIDSTLSTTSSNKLAASVIIWQD 359
Query: 563 PQNNSTYLKIKIANIGS 579
+ YL+IK N G+
Sbjct: 360 SSDKRNYLRIKAVNFGA 410
>TC204589 similar to UP|Q76LU4 (Q76LU4) Alpha-L-arabinofuranosidase, partial
(14%)
Length = 808
Score = 99.8 bits (247), Expect = 3e-21
Identities = 71/170 (41%), Positives = 91/170 (52%), Gaps = 11/170 (6%)
Frame = +3
Query: 1 MTFSKACCSFLWLYLIIVSFVAFQCNA---NGSQISSLVVNA--AQGRPMPNTLFGIFYE 55
M SKA CS L+L IV + FQC A + ++L VNA A G+P+P TLFGIF+E
Sbjct: 222 MGSSKALCSVSLLHLFIVVCLVFQCFATKKHADLTTTLTVNASDASGKPIPETLFGIFFE 401
Query: 56 EINHAGSGGIWAQLVNNSGFEAAGTRTPSNIFPWTIIGTESSVKLQTELSSCFERNKVAL 115
EINHAG+GG+WA+LV+N GFEA G TPSNI ++ L+ R +V
Sbjct: 402 EINHAGAGGLWAELVSNRGFEAGGPNTPSNIALGQLLEMNH**ILKLIAPLALTRTRV-- 575
Query: 116 RMDVLC------DKCPPDGVGVSNPGFWGMNIVQGKKYKVVFFYRSLGSL 159
D C P DGV V+ GMNI Q ++ F+ R L L
Sbjct: 576 HSDWRCM*HQX*HIWPADGV-VAITLVLGMNIEQXEEXXXGFYCRQLDHL 722
>TC213724 weakly similar to UP|Q944F3 (Q944F3) Arabinosidase ARA-1, partial
(10%)
Length = 595
Score = 97.1 bits (240), Expect = 2e-20
Identities = 47/67 (70%), Positives = 55/67 (81%)
Frame = +3
Query: 508 WNPDAIVFNSYQAYGTPSYWVTYMFKESNGATFLNSQLQTPDPGSLIASAILCQSPQNNS 567
W+PDAIVFNS Q Y TPSY V +MF+ESNGATFL SQLQTPDP S+ ASAIL ++ Q+
Sbjct: 144 WSPDAIVFNSNQVYETPSYRVQFMFRESNGATFLKSQLQTPDPDSVDASAILWKNLQDKK 323
Query: 568 TYLKIKI 574
TYLKIK+
Sbjct: 324 TYLKIKV 344
>TC204591 homologue to UP|Q6RIB6 (Q6RIB6) Cytosolic malate dehydrogenase ,
partial (84%)
Length = 1604
Score = 83.2 bits (204), Expect = 3e-16
Identities = 43/70 (61%), Positives = 53/70 (75%)
Frame = +2
Query: 416 PLNHPADLYDYHTYPIDARAMFNAYHDFDKSPRNGPKAFVSEYALIGALQAKYGTLLGAV 475
PL+HPAD+YDYH Y +A MF+ + FD + RNGPKAFVSEYA+ G A G+LL A+
Sbjct: 1400 PLDHPADMYDYHVYT-NANDMFSRANAFDHTSRNGPKAFVSEYAVTGK-DAGTGSLLAAL 1573
Query: 476 SEAGFLIGLE 485
+EAGFLIGLE
Sbjct: 1574 AEAGFLIGLE 1603
>TC212850 weakly similar to UP|Q76LU3 (Q76LU3) Alpha-L-arabinofuranosidase,
partial (3%)
Length = 558
Score = 67.0 bits (162), Expect = 2e-11
Identities = 41/108 (37%), Positives = 53/108 (48%), Gaps = 35/108 (32%)
Frame = +2
Query: 577 IGSIPVNLKISLQGYVSKNLAGSTKTVLTSGNILDENTFAAP------------------ 618
+GS +N KISL G+ S N +TKTVLTS N LDEN+ A P
Sbjct: 8 VGSNQINFKISLNGFESSNATKATKTVLTSKNALDENSLANPTNVHITSFILFLLL*TNN 187
Query: 619 -----------------KKIAPQTSPLQNPGNEMNVMIPPVSLTVLDM 649
+I P+ SPLQ+ NE+N ++PPVSLTV D+
Sbjct: 188 TYNN*LS*HLMFICLFCTQIVPKRSPLQSANNEINDILPPVSLTVFDL 331
>TC208756 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(5%)
Length = 544
Score = 57.8 bits (138), Expect = 2e-08
Identities = 27/51 (52%), Positives = 36/51 (69%)
Frame = +1
Query: 598 GSTKTVLTSGNILDENTFAAPKKIAPQTSPLQNPGNEMNVMIPPVSLTVLD 648
GST TVLTS N++DEN+F PKK+ PQTSPL+ + V++P S+ LD
Sbjct: 22 GSTITVLTSTNVMDENSFLEPKKVVPQTSPLEIDDKHLIVLLPSYSVISLD 174
>TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-related
protein, partial (5%)
Length = 647
Score = 34.7 bits (78), Expect = 0.14
Identities = 16/38 (42%), Positives = 28/38 (73%)
Frame = +3
Query: 140 NIVQGKKYKVVFFYRSLGSLDMRVAFRDAISGRILASS 177
NI +G+KYKVVF+ ++ G++++ V+F + G LAS+
Sbjct: 129 NIEKGQKYKVVFYAKADGAINLDVSFVGSEKGEKLASN 242
>TC210739 weakly similar to UP|Q8S9K7 (Q8S9K7) At1g48640/F11I4_17, partial
(19%)
Length = 926
Score = 28.5 bits (62), Expect = 9.8
Identities = 9/21 (42%), Positives = 14/21 (65%)
Frame = +1
Query: 514 VFNSYQAYGTPSYWVTYMFKE 534
+ NS+ + GTP+ W+ YM E
Sbjct: 211 ILNSFMSMGTPNSWIKYMIPE 273
>CD398081
Length = 513
Score = 28.5 bits (62), Expect = 9.8
Identities = 24/81 (29%), Positives = 41/81 (49%)
Frame = -3
Query: 480 FLIGLERNSDHVAMASYAPLLVNANDRNWNPDAIVFNSYQAYGTPSYWVTYMFKESNGAT 539
FL+G++ + + +A+ + LLV DR++ +V +++Q Y S+ + ES G
Sbjct: 457 FLVGVDDDENELAILEFIHLLVETMDRHFG--NVVSSAFQTYLFSSF--PF*NVESRG-- 296
Query: 540 FLNSQLQTPDPGSLIASAILC 560
S L P + AS ILC
Sbjct: 295 ---SFLYVSSPYTCSAS*ILC 242
>CA801925
Length = 457
Score = 28.5 bits (62), Expect = 9.8
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = +1
Query: 7 CCSFLWLYLIIVSFVAFQC 25
CC FL +YL+ VS ++F C
Sbjct: 202 CCVFLIIYLLSVSVISFNC 258
>TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(78%)
Length = 1607
Score = 28.5 bits (62), Expect = 9.8
Identities = 19/54 (35%), Positives = 26/54 (47%), Gaps = 1/54 (1%)
Frame = +3
Query: 209 NLTLTTTKEGTVWLDQVSAMPTDTFKGHGFRKDLVEMLIQLK-PAFLRFPGGCF 261
N +L K GT + V + D+F+GH R V L QL P ++ G CF
Sbjct: 408 NQSLAAAKPGTGVVVAVKRLSLDSFQGHKDRLAEVNYLGQLSHPHLVKLIGYCF 569
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,102,284
Number of Sequences: 63676
Number of extensions: 451556
Number of successful extensions: 2091
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 2075
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2082
length of query: 650
length of database: 12,639,632
effective HSP length: 103
effective length of query: 547
effective length of database: 6,081,004
effective search space: 3326309188
effective search space used: 3326309188
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC141107.11


