

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.2 + phase: 0
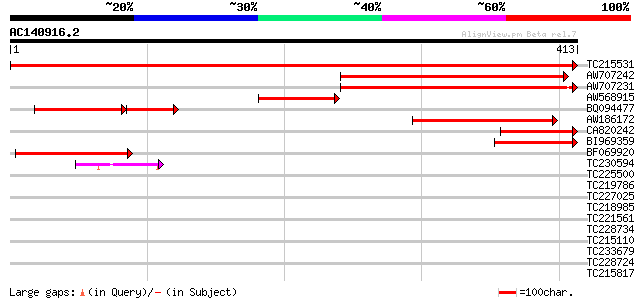
(413 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC215531 UP|HEM2_SOYBN (P43210) Delta-aminolevulinic acid dehydr... 722 0.0
AW707242 similar to SP|P43210|HEM2 Delta-aminolevulinic acid deh... 280 1e-75
AW707231 weakly similar to SP|P43210|HEM2 Delta-aminolevulinic a... 207 5e-54
AW568915 SP|P43210|HEM2 Delta-aminolevulinic acid dehydratase c... 123 2e-28
BQ094477 SP|P43210|HEM2 Delta-aminolevulinic acid dehydratase c... 76 3e-28
AW186172 similar to SP|P30124|HEM2_ Delta-aminolevulinic acid de... 113 1e-25
CA820242 SP|P43210|HEM2 Delta-aminolevulinic acid dehydratase c... 105 4e-23
BI969359 similar to SP|P43210|HEM2_ Delta-aminolevulinic acid de... 103 2e-22
BF069920 homologue to SP|P43210|HEM2_ Delta-aminolevulinic acid ... 90 2e-18
TC230594 homologue to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP... 41 9e-04
TC225500 homologue to UP|Q6RJY1 (Q6RJY1) 60S ribosomal protein L... 35 0.062
TC219786 similar to GB|AAM91354.1|22137018|AY133524 At2g28390/T1... 33 0.18
TC227025 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate d... 33 0.31
TC218985 similar to UP|HPPD_DAUCA (O23920) 4-hydroxyphenylpyruva... 33 0.31
TC221561 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate d... 32 0.40
TC228734 weakly similar to UP|Q99738 (Q99738) Pinin, partial (4%) 32 0.40
TC215110 similar to UP|Q9SKU1 (Q9SKU1) Expressed protein (At2g20... 32 0.52
TC233679 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 32 0.52
TC228724 similar to UP|Q9FML2 (Q9FML2) Histone deacetylase, part... 32 0.52
TC215817 similar to GB|AAM78057.1|21928057|AY125547 AT4g34180/F2... 32 0.52
>TC215531 UP|HEM2_SOYBN (P43210) Delta-aminolevulinic acid dehydratase,
chloroplast precursor (Porphobilinogen synthase)
(ALADH) , complete
Length = 1737
Score = 722 bits (1864), Expect = 0.0
Identities = 362/414 (87%), Positives = 382/414 (91%), Gaps = 1/414 (0%)
Frame = +2
Query: 1 MASSSTIPNTPLTLNSHTYVDLKPALPLKNYLSFSSSK-RRPPCLFTVRASDADFEAAVV 59
MA +S+IPN P NS +YV L+ L N+ S ++K R LF VRASD++FEAAVV
Sbjct: 155 MAMASSIPNAPSAFNSQSYVGLRAPLRTFNFSSPQAAKIPRSQRLFVVRASDSEFEAAVV 334
Query: 60 AGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANFVYPLF 119
AG VP APPVPP PAAP GTPVV SLP+ RRPRRNR+S RSAFQET++SPANFVYPLF
Sbjct: 335 AGKVPPAPPVPPRPAAPVGTPVVPSLPLHRRPRRNRKSPALRSAFQETSISPANFVYPLF 514
Query: 120 IHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYN 179
IHEGEEDTPIGAMPGCYRLGWRHGL+EEVAKARDVGVNSVVLFPKIPDALK+PTGDEAYN
Sbjct: 515 IHEGEEDTPIGAMPGCYRLGWRHGLVEEVAKARDVGVNSVVLFPKIPDALKSPTGDEAYN 694
Query: 180 ENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVA 239
ENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVA
Sbjct: 695 ENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVA 874
Query: 240 QARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPR 299
QA+AGADVVSPSDMMDGRVGA+RAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPR
Sbjct: 875 QAQAGADVVSPSDMMDGRVGALRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPR 1054
Query: 300 FGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQV 359
FGDKKTYQMNPANYREALTEMREDE+EGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQV
Sbjct: 1055FGDKKTYQMNPANYREALTEMREDESEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQV 1234
Query: 360 SGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEKR 413
SGEY+MIKA GALKMIDEEKVMMESL+CLRRAGADIILTY ALQAARCLCGEKR
Sbjct: 1235SGEYAMIKAAGALKMIDEEKVMMESLMCLRRAGADIILTYSALQAARCLCGEKR 1396
>AW707242 similar to SP|P43210|HEM2 Delta-aminolevulinic acid dehydratase
chloroplast precursor (EC 4.2.1.24), partial (40%)
Length = 508
Score = 280 bits (715), Expect = 1e-75
Identities = 139/166 (83%), Positives = 152/166 (90%)
Frame = +2
Query: 242 RAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFG 301
+AGA+VVSP DMMDGRVG + A LDAEGFQHVSIM+Y+AKYASSFYGPFRE L+SNPRFG
Sbjct: 2 QAGANVVSPCDMMDGRVGTLGATLDAEGFQHVSIMAYSAKYASSFYGPFREGLNSNPRFG 181
Query: 302 DKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSG 361
DKKTY MNPA YR+ALTEMREDE+EG DILLVKPGLPYLDIIRL+RDNSPLPIAAYQ SG
Sbjct: 182 DKKTY*MNPAYYRKALTEMREDESEGTDILLVKPGLPYLDIIRLVRDNSPLPIAAYQGSG 361
Query: 362 EYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARC 407
EY+MIKAGGA KMID+EK MME+LLCLRRAGADI+L Y ALQA RC
Sbjct: 362 EYAMIKAGGASKMIDQEKGMMEALLCLRRAGADIMLAYSALQAGRC 499
>AW707231 weakly similar to SP|P43210|HEM2 Delta-aminolevulinic acid
dehydratase chloroplast precursor (EC 4.2.1.24),
partial (39%)
Length = 514
Score = 207 bits (528), Expect = 5e-54
Identities = 108/172 (62%), Positives = 133/172 (76%)
Frame = +2
Query: 242 RAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFG 301
+AG D P D MD RVG +R+ LDA FQHVSIM+Y+AKYA+SFYGPFR+AL+SNPR G
Sbjct: 2 QAGTDGAKPYDFMDCRVGTLRSTLDA*RFQHVSIMAYSAKYANSFYGPFRKALNSNPRVG 181
Query: 302 DKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSG 361
D+KTY +NPA YR+A TEM +DE+ G DILLVKPG PYLD IRLL+D LPIAAYQ S
Sbjct: 182 DEKTYSINPAYYRKAFTEMPDDES*GTDILLVKPGPPYLDFIRLLKDYFALPIAAYQGSW 361
Query: 362 EYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEKR 413
+ + IKAG AL+MI E+ MME L+CL+ AGADI L Y A+Q A+ +C E++
Sbjct: 362 DIAKIKAGCALQMIHEKNGMMEVLMCLQMAGADISLPYSAVQTAK-VCVERK 514
>AW568915 SP|P43210|HEM2 Delta-aminolevulinic acid dehydratase chloroplast
precursor (EC 4.2.1.24), partial (14%)
Length = 347
Score = 123 bits (308), Expect = 2e-28
Identities = 59/59 (100%), Positives = 59/59 (100%)
Frame = +1
Query: 182 GLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQ 240
GLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQ
Sbjct: 7 GLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQ 183
>BQ094477 SP|P43210|HEM2 Delta-aminolevulinic acid dehydratase chloroplast
precursor (EC 4.2.1.24), partial (25%)
Length = 422
Score = 75.9 bits (185), Expect(2) = 3e-28
Identities = 42/68 (61%), Positives = 48/68 (69%), Gaps = 1/68 (1%)
Frame = +2
Query: 19 YVDLKPALPLKNYLSFSSSK-RRPPCLFTVRASDADFEAAVVAGNVPDAPPVPPTPAAPA 77
YV L+ L N+ S ++K R LF VRASD++FEAAVVAG VP APPVPP PAAP
Sbjct: 2 YVGLRAPLRTFNFSSPQAAKIPRSQRLFVVRASDSEFEAAVVAGKVPPAPPVPPRPAAPV 181
Query: 78 GTPVVTSL 85
GTPVV SL
Sbjct: 182GTPVVPSL 205
Score = 67.8 bits (164), Expect(2) = 3e-28
Identities = 30/38 (78%), Positives = 34/38 (88%)
Frame = +3
Query: 86 PIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIHEG 123
P+ RRPRRNR+S RSAFQET++SPANFVYPLFIHEG
Sbjct: 306 PLHRRPRRNRKSPALRSAFQETSISPANFVYPLFIHEG 419
>AW186172 similar to SP|P30124|HEM2_ Delta-aminolevulinic acid dehydratase
chloroplast precursor (EC 4.2.1.24), partial (18%)
Length = 327
Score = 113 bits (283), Expect = 1e-25
Identities = 66/108 (61%), Positives = 79/108 (73%), Gaps = 2/108 (1%)
Frame = +2
Query: 294 LDSNP-RFGDKKTYQMNPAN-YREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSP 351
L+SNP + KKT +M P + LTEMREDE+EGADILLV PGL YL I+ LLRDNSP
Sbjct: 2 LNSNPQKEKTKKTNKMKPN**QKRHLTEMREDESEGADILLVGPGLSYLHILTLLRDNSP 181
Query: 352 LPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTY 399
LPIAAY VSG ++I A GA+ ++D V+MESLL L AGAD+ LTY
Sbjct: 182 LPIAAYHVSGGAALINAAGAITLVDHHFVVMESLLFLPWAGADMGLTY 325
>CA820242 SP|P43210|HEM2 Delta-aminolevulinic acid dehydratase chloroplast
precursor (EC 4.2.1.24), partial (13%)
Length = 404
Score = 105 bits (262), Expect = 4e-23
Identities = 52/56 (92%), Positives = 54/56 (95%)
Frame = -2
Query: 358 QVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEKR 413
QVSGEY+MIKA GALKMIDEEKVMMESL+CLRRAGADIILTY ALQAARCLCGEKR
Sbjct: 367 QVSGEYAMIKAAGALKMIDEEKVMMESLMCLRRAGADIILTYSALQAARCLCGEKR 200
>BI969359 similar to SP|P43210|HEM2_ Delta-aminolevulinic acid dehydratase
chloroplast precursor (EC 4.2.1.24), partial (14%)
Length = 430
Score = 103 bits (256), Expect = 2e-22
Identities = 50/60 (83%), Positives = 54/60 (89%)
Frame = -3
Query: 354 IAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEKR 413
IAA V GEY+MIKA GALKMIDEEKVMMES +CLRR GADIILTY+ALQA+RCLCGEKR
Sbjct: 428 IAAXXVXGEYAMIKAAGALKMIDEEKVMMESXMCLRRTGADIILTYYALQASRCLCGEKR 249
>BF069920 homologue to SP|P43210|HEM2_ Delta-aminolevulinic acid
dehydratase chloroplast precursor (EC 4.2.1.24),
partial (20%)
Length = 362
Score = 90.1 bits (222), Expect = 2e-18
Identities = 48/86 (55%), Positives = 58/86 (66%), Gaps = 1/86 (1%)
Frame = +1
Query: 5 STIPNTPLTLNSHTYVDLKPALPLKNYLSFSSSK-RRPPCLFTVRASDADFEAAVVAGNV 63
++IPN P NS +Y L+ L N+ S ++K R LF VRASD++FEAAVVAG V
Sbjct: 1 ASIPNAPSAFNSQSYFGLRAPLRTFNFSSPQAAKIPRSQRLFVVRASDSEFEAAVVAGKV 180
Query: 64 PDAPPVPPTPAAPAGTPVVTSLPIQR 89
P APPVPP PAA GTPVV SLP+ R
Sbjct: 181PXAPPVPPKPAASVGTPVVASLPLHR 258
>TC230594 homologue to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP4-like,
partial (24%)
Length = 928
Score = 41.2 bits (95), Expect = 9e-04
Identities = 28/75 (37%), Positives = 35/75 (46%), Gaps = 11/75 (14%)
Frame = +1
Query: 49 ASDADFEAAVVAGNV-----PDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSA 103
AS A A VAGN P PP PP P PA T +T+LP+ R+P R H +
Sbjct: 439 ASSATSPLAAVAGNQNAPTSPKPPPQPPPP--PAATITITTLPLPRKPLRQPHQRHHNNR 612
Query: 104 FQE------TTLSPA 112
+ TL+PA
Sbjct: 613 LRHLS*SIILTLTPA 657
>TC225500 homologue to UP|Q6RJY1 (Q6RJY1) 60S ribosomal protein L12, complete
Length = 1130
Score = 35.0 bits (79), Expect = 0.062
Identities = 23/77 (29%), Positives = 37/77 (47%)
Frame = +1
Query: 33 SFSSSKRRPPCLFTVRASDADFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPR 92
S +S ++PP AS + +V +PP PP+ + P+ +P T+ RRPR
Sbjct: 409 SEKTSPKKPPRTGRASASPSSSPFRIVRRRSRWSPPPPPSLSRPSRSPSATA----RRPR 576
Query: 93 RNRRSATHRSAFQETTL 109
+ +AT RS +L
Sbjct: 577 TSSTTATSRSMTSSRSL 627
>TC219786 similar to GB|AAM91354.1|22137018|AY133524 At2g28390/T1B3.9
{Arabidopsis thaliana;} , partial (32%)
Length = 848
Score = 33.5 bits (75), Expect = 0.18
Identities = 22/59 (37%), Positives = 27/59 (45%), Gaps = 2/59 (3%)
Frame = +3
Query: 43 CLFTVRASDADFEAAVVAGNVPDAPPVPPTPAAPAGTPV--VTSLPIQRRPRRNRRSAT 99
CL + RAS + + PD PP PP P A A TP+ + P Q PR S T
Sbjct: 93 CLLSHRASSIPNHRSSLTDPPPDPPPTPPVPPA-AVTPLKGAAAAPPQASPRSMTSSTT 266
>TC227025 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate
dehydrogenase, complete
Length = 2097
Score = 32.7 bits (73), Expect = 0.31
Identities = 17/34 (50%), Positives = 21/34 (61%)
Frame = +2
Query: 64 PDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRS 97
P PP PP+ A+P+ TP TS P RRP +RS
Sbjct: 356 PWPPPWPPSAASPSSTP--TSPPPSRRPSSAKRS 451
>TC218985 similar to UP|HPPD_DAUCA (O23920) 4-hydroxyphenylpyruvate
dioxygenase (4HPPD) (HPD) (HPPDase) , partial (35%)
Length = 598
Score = 32.7 bits (73), Expect = 0.31
Identities = 34/111 (30%), Positives = 48/111 (42%), Gaps = 2/111 (1%)
Frame = +2
Query: 3 SSSTIPNTPLTLNSHTYVDLKPALPLKNYLSFSSSKRRPPCLFTVRASDADFEAAVVAGN 62
SSS P P T + + DL+ L +N +S +K PP + A+ +
Sbjct: 149 SSSGAPMPP-TPPADSLGDLE-CLLWQNLISPPETKSTPPTS-SAPATSPSSSPLLTLPL 319
Query: 63 VPDAPPVPPTPAAPAGT--PVVTSLPIQRRPRRNRRSATHRSAFQETTLSP 111
P APP+PP P P T P + SLP N SA+ S ++ T P
Sbjct: 320 SPPAPPLPPPPPFPVSTPPPALPSLP-------NTASASAPSPWKSPTRKP 451
>TC221561 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate
dehydrogenase, complete
Length = 2001
Score = 32.3 bits (72), Expect = 0.40
Identities = 29/90 (32%), Positives = 40/90 (44%), Gaps = 8/90 (8%)
Frame = +2
Query: 16 SHTYVDLKPALPLKNYLSFSSS--------KRRPPCLFTVRASDADFEAAVVAGNVPDAP 67
S+TY D+ + L +Y+ F++ RR P AS D P
Sbjct: 251 SYTYDDV---IFLPHYIDFAADVVDLSTRLTRRLPLAVPFVASPMD-----TVSESAWPP 406
Query: 68 PVPPTPAAPAGTPVVTSLPIQRRPRRNRRS 97
P PP+ A+P+ TP TS P RRP +RS
Sbjct: 407 PWPPSAASPSSTP--TSPPPSRRPSSAKRS 490
>TC228734 weakly similar to UP|Q99738 (Q99738) Pinin, partial (4%)
Length = 1156
Score = 32.3 bits (72), Expect = 0.40
Identities = 31/108 (28%), Positives = 42/108 (38%), Gaps = 11/108 (10%)
Frame = +1
Query: 2 ASSSTIPNTPLTLNSHTYVDLKPALPLKNYLSFSSSKRRPPCLFTVRASDADFEAA---- 57
+SS T P+TP T + P PL + S S R P + + ++F
Sbjct: 355 SSSKTPPSTPST----SPPPATPLPPLPQPPTPSPSPCRSPSPPSTPTTASEFTTPNSTP 522
Query: 58 ---VVAGNVPDAPPVPP----TPAAPAGTPVVTSLPIQRRPRRNRRSA 98
A P PP PP T P+G P T P P R+R S+
Sbjct: 523 TPPTAASRSPSPPPSPPPTRATATPPSGPPTSTPPPSPSPPSRSRSSS 666
>TC215110 similar to UP|Q9SKU1 (Q9SKU1) Expressed protein
(At2g20760/F5H14.27), partial (47%)
Length = 1225
Score = 32.0 bits (71), Expect = 0.52
Identities = 20/51 (39%), Positives = 24/51 (46%), Gaps = 4/51 (7%)
Frame = +3
Query: 67 PPVPPTPAAPAGT----PVVTSLPIQRRPRRNRRSATHRSAFQETTLSPAN 113
PP PPTP+ T P TS I PRR RRS+ Q T P++
Sbjct: 126 PPPPPTPSLATPTSPPSPPTTSPSITPTPRRRRRSSDSAIPVQFTPSLPSS 278
>TC233679 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (7%)
Length = 922
Score = 32.0 bits (71), Expect = 0.52
Identities = 17/43 (39%), Positives = 22/43 (50%)
Frame = +3
Query: 64 PDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQE 106
P +PP PPTPA P+ P P + PR++ S HR E
Sbjct: 93 PSSPPSPPTPAPPSPAP---ETPEKIPPRQS*ASEHHRRCRHE 212
>TC228724 similar to UP|Q9FML2 (Q9FML2) Histone deacetylase, partial (82%)
Length = 1677
Score = 32.0 bits (71), Expect = 0.52
Identities = 30/104 (28%), Positives = 43/104 (40%), Gaps = 3/104 (2%)
Frame = +2
Query: 3 SSSTIPNTPLTLNSHTYVDLKPALPLKNYLSFSSSKRRPPCLFTVRASDADFEAAVVAGN 62
S+++ P T T + + P P S S+S R P +S A +A
Sbjct: 287 SAASTPTTTSTSSPPSPPRPSPIPPSLVTSSASTSARTAP------SSTASSPSARPPPA 448
Query: 63 VPDAPP---VPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSA 103
P APP PTP +P+ P + +P RP + S T SA
Sbjct: 449 APSAPPSNSTAPTPTSPSIGPAASIMPRSLRPLDSATSTTLSSA 580
>TC215817 similar to GB|AAM78057.1|21928057|AY125547 AT4g34180/F28A23_60
{Arabidopsis thaliana;} , partial (67%)
Length = 1233
Score = 32.0 bits (71), Expect = 0.52
Identities = 20/53 (37%), Positives = 23/53 (42%), Gaps = 12/53 (22%)
Frame = -3
Query: 61 GNVPDAPPVPPTPA------------APAGTPVVTSLPIQRRPRRNRRSATHR 101
G+V APP PTPA AP + P RRPR R +THR
Sbjct: 304 GSVARAPPSSPTPASRGHICE*CRRFAPPRILLAVEEPEHRRPRPRRGGSTHR 146
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,411,793
Number of Sequences: 63676
Number of extensions: 233640
Number of successful extensions: 3215
Number of sequences better than 10.0: 133
Number of HSP's better than 10.0 without gapping: 2364
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2886
length of query: 413
length of database: 12,639,632
effective HSP length: 100
effective length of query: 313
effective length of database: 6,272,032
effective search space: 1963146016
effective search space used: 1963146016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC140916.2


