

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.6 - phase: 0
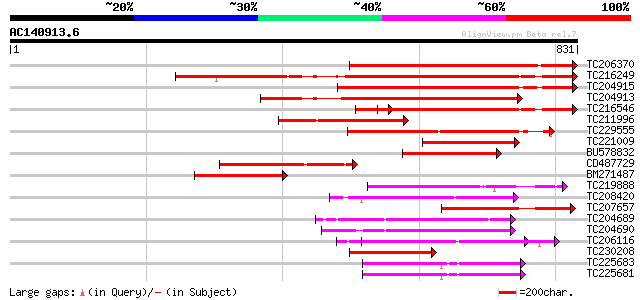
(831 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, part... 551 e-157
TC216249 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 500 e-141
TC204915 weakly similar to GB|AAH46286.1|28279482|BC046286 spast... 416 e-116
TC204913 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 381 e-106
TC216546 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 345 5e-95
TC211996 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, part... 298 5e-81
TC229555 similar to GB|AAM62497.1|21553404|AY085265 26S proteaso... 258 6e-69
TC221009 homologue to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, pa... 251 9e-67
BU578832 237 1e-62
CD487729 similar to PIR|T02901|T02 MSP1 protein homolog T13J8.11... 233 3e-61
BM271487 similar to GP|15810167|gb At1g64110/F22C12_22 {Arabidop... 224 2e-58
TC219888 similar to UP|Q944N4 (Q944N4) Tobacco mosaic virus heli... 211 1e-54
TC208420 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (79%) 202 5e-52
TC207657 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic ... 199 4e-51
TC204689 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type A... 192 7e-49
TC204690 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type A... 192 7e-49
TC206116 UP|CC48_SOYBN (P54774) Cell division cycle protein 48 h... 186 5e-47
TC230208 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic ... 170 2e-42
TC225683 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit ... 155 9e-38
TC225681 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit ... 155 9e-38
>TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (38%)
Length = 1502
Score = 551 bits (1419), Expect = e-157
Identities = 282/334 (84%), Positives = 310/334 (92%), Gaps = 1/334 (0%)
Frame = +2
Query: 499 EFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILL 558
EFEKRIRPEVIPANEIGVTF+DIGALDE K+SLQELVMLPLRRPDLF+GGLLKPCRGILL
Sbjct: 2 EFEKRIRPEVIPANEIGVTFADIGALDEIKESLQELVMLPLRRPDLFKGGLLKPCRGILL 181
Query: 559 FGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFV 618
FGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKV+PTIIFV
Sbjct: 182 FGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVAPTIIFV 361
Query: 619 DEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRR 678
DEVDSMLGQRTRVGEHEAMRKIKNEFM++WDGL + ++ILVLAATNRPFDLDEAIIRR
Sbjct: 362 DEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLLTGPNEQILVLAATNRPFDLDEAIIRR 541
Query: 679 FERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRP 738
FERRI+VGLPS ENRE IL+TLLAKEK H LDFKELATMTEGY+GSDLKNLC TAAYRP
Sbjct: 542 FERRILVGLPSVENREMILKTLLAKEK-HENLDFKELATMTEGYTGSDLKNLCITAAYRP 718
Query: 739 VRELIQQEIQKD-SKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAA 797
VRELIQQE KD KKK++AEGQ+S+DA + K++ E+E ITLRPLNM+D + AK+QVAA
Sbjct: 719 VRELIQQERMKDMEKKKREAEGQSSEDASNNKDKEEKE--ITLRPLNMEDMRQAKTQVAA 892
Query: 798 SFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
SFA+EG+ MNEL+ WNDLYGEGGSRKK+QL+YFL
Sbjct: 893 SFASEGSVMNELKHWNDLYGEGGSRKKQQLTYFL 994
>TC216249 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial
(14%)
Length = 2222
Score = 500 bits (1287), Expect = e-141
Identities = 278/627 (44%), Positives = 393/627 (62%), Gaps = 39/627 (6%)
Frame = +2
Query: 244 EKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILIIGS-- 301
+K+LI +L++V+ S++ P +L+M+DA+K + + + F++ L L +++IGS
Sbjct: 182 DKLLIHSLFEVVFSESRSAPFILFMKDAEKSIVGNGDSHS-FKSKLENLPDNVVVIGSHT 358
Query: 302 ------------------------------------RILDSGNECKRVDEMLTSLFPYNI 325
R+ D G E + + LT LFP I
Sbjct: 359 QNDSRKEKSHPGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEAPKQNRTLTKLFPNKI 538
Query: 326 EIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLS 385
I P+DE+ L SWK Q + D++ ++I+ N +H+ VL ++C L+++C+ D + +
Sbjct: 539 TIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLGRCGMECEGLETLCIKDQTLTN 718
Query: 386 NYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAV 445
E+II A+S+H+M+N E + + KL++ C S+ + +GI Q ++
Sbjct: 719 ENAEKIIGWALSHHLMQNSEAK-PDSKLVLSCESILYGIGILQ---------------SI 850
Query: 446 TSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIR 505
+E K S++K+ K V +NEFEKR+
Sbjct: 851 QNESK---------------------------SLKKS--------LKDVVTENEFEKRLL 925
Query: 506 PEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGT 564
+VIP ++I VTF DIGAL++ KD+L+ELVMLPL+RP+LF +G L KPC+GILLFGPPGT
Sbjct: 926 ADVIPPSDIDVTFDDIGALEKVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGT 1105
Query: 565 GKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSM 624
GKTMLAKAIA EAGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K+SP++IFVDEVDSM
Sbjct: 1106GKTMLAKAIATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKISPSVIFVDEVDSM 1285
Query: 625 LGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIM 684
LG+R GEHEAMRK+KNEFM NWDGL +K +R+LVLAATNRPFDLDEA+IRR RR+M
Sbjct: 1286LGRRENPGEHEAMRKMKNEFMVNWDGLRTKETERVLVLAATNRPFDLDEAVIRRMPRRLM 1465
Query: 685 VGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQ 744
V LP A NR IL+ +LAKE++ +D +A+MT+GYSGSDLKNLC TAA+RP++E+++
Sbjct: 1466VNLPDAPNRAKILKVILAKEELSPDVDLDAVASMTDGYSGSDLKNLCVTAAHRPIKEILE 1645
Query: 745 QEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGA 804
+E K + AEGQ + + + +R LNM+DFK A QV AS ++E
Sbjct: 1646KE--KKERAAALAEGQPAPALCSSGD---------VRSLNMEDFKYAHQQVCASVSSESV 1792
Query: 805 GMNELRQWNDLYGEGGSRKKEQLSYFL 831
M EL QWN+LYGEGGSR K+ LSYF+
Sbjct: 1793NMTELLQWNELYGEGGSRVKKALSYFM 1873
>TC204915 weakly similar to GB|AAH46286.1|28279482|BC046286 spastic
paraplegia 4 homolog {Mus musculus;} , partial (21%)
Length = 1470
Score = 416 bits (1070), Expect = e-116
Identities = 210/352 (59%), Positives = 267/352 (75%), Gaps = 1/352 (0%)
Frame = +1
Query: 481 KTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLR 540
+ + +N + K V +NEFEK++ +VIP +IGVTF DIGAL+ KD+L+ELVMLPL+
Sbjct: 166 QNENKNLKKSLKDVVTENEFEKKLLADVIPPTDIGVTFDDIGALENVKDTLKELVMLPLQ 345
Query: 541 RPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEK 599
RP+LF +G L KPC+GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE EK
Sbjct: 346 RPELFCKGQLAKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEK 525
Query: 600 NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRI 659
V+A+F+LA+K++P++IFVDEVDSMLG+R EHEAMRK+KNEFM NWDGL +K ++R+
Sbjct: 526 YVKAVFSLASKIAPSVIFVDEVDSMLGRRENPSEHEAMRKMKNEFMVNWDGLRTKDKERV 705
Query: 660 LVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMT 719
LVLAATNRPFDLDEA+IRR RR+MV LP A NRE ILR +L KE + +DF+ +A MT
Sbjct: 706 LVLAATNRPFDLDEAVIRRLPRRLMVNLPDAPNREKILRVILVKEDLAPDVDFEAIANMT 885
Query: 720 EGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVIT 779
+GYSGSDLKNLC TAA+ P+RE++++E KK++ S+ +
Sbjct: 886 DGYSGSDLKNLCVTAAHCPIREILEKE-----KKERSLALSESKPLPGLCGSGD------ 1032
Query: 780 LRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
+RPL M DF+ A QV AS ++E MNEL QWNDLYGEGGSRK LSYF+
Sbjct: 1033IRPLKMDDFRYAHEQVCASVSSESTNMNELLQWNDLYGEGGSRKMRSLSYFM 1188
>TC204913 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial
(14%)
Length = 1080
Score = 381 bits (978), Expect = e-106
Identities = 200/385 (51%), Positives = 263/385 (67%), Gaps = 1/385 (0%)
Frame = +3
Query: 368 LDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIF 427
LDC DL+++C+ D + + +E+II AISYH M + E R+ KL+I S+ + I
Sbjct: 6 LDCPDLETLCIKDHTLTTESVEKIIGWAISYHFMHSSEASIRDSKLVISAESIKYGHKIL 185
Query: 428 QAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENS 487
Q + +E K N
Sbjct: 186 QG---------------IQNENK-----------------------------------NM 215
Query: 488 VPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLF-E 546
+ K V +NEFEK++ +VIP +IGVTF DIGAL+ K++L+ELVMLPL+RP+LF +
Sbjct: 216 KKSLKDVVTENEFEKKLLTDVIPPTDIGVTFDDIGALENVKETLKELVMLPLQRPELFGK 395
Query: 547 GGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFT 606
G L KPC+GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE EK V+A+F+
Sbjct: 396 GQLAKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFS 575
Query: 607 LAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATN 666
LA+K++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM NWDGL +K ++RILVLAATN
Sbjct: 576 LASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDKERILVLAATN 755
Query: 667 RPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSD 726
RPFDLDEA+IRR RR+MV LP A NR I+R +LAKE + +DF+ +A MT+GYSGSD
Sbjct: 756 RPFDLDEAVIRRLPRRLMVNLPDAPNRGKIVRVILAKEDLAPDVDFEAIANMTDGYSGSD 935
Query: 727 LKNLCTTAAYRPVRELIQQEIQKDS 751
LKNLC TAA P+R+++++E ++ S
Sbjct: 936 LKNLCVTAAQCPIRQILEKEKKERS 1010
>TC216546 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial
(14%)
Length = 1321
Score = 345 bits (885), Expect = 5e-95
Identities = 173/293 (59%), Positives = 222/293 (75%), Gaps = 1/293 (0%)
Frame = +3
Query: 540 RRPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDE 598
R P F +G PC+GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE E
Sbjct: 99 RGPSYFAKGN*PXPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGE 278
Query: 599 KNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDR 658
K V+A+F+LA+K++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM NWDGL +K +R
Sbjct: 279 KYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTER 458
Query: 659 ILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATM 718
+LVLAATNRPFDLDEA+IRR RR+MV LP A NR IL+ +L KE + +D +A+M
Sbjct: 459 VLVLAATNRPFDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILEKEDLSSDIDMDAIASM 638
Query: 719 TEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVI 778
T+GYSGSDLKNLC TAA+RP++E++++E K + +EG+ + + +
Sbjct: 639 TDGYSGSDLKNLCVTAAHRPIKEILEKE--KKEQAAAVSEGRPAPALSGSGD-------- 788
Query: 779 TLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
+R LNM+DFK A QV AS ++E M EL+QWN+LYGEGGSR K+ LSYF+
Sbjct: 789 -IRSLNMEDFKYAHQQVCASVSSESINMTELQQWNELYGEGGSRVKKALSYFM 944
Score = 64.3 bits (155), Expect = 2e-10
Identities = 33/56 (58%), Positives = 42/56 (74%), Gaps = 1/56 (1%)
Frame = +2
Query: 508 VIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPP 562
VIP N+IGVTF DIGAL+ KD+L+ELVMLPL+RP+LF +G L K +G + P
Sbjct: 2 VIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKXVQGYPIIWSP 169
>TC211996 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (17%)
Length = 580
Score = 298 bits (764), Expect = 5e-81
Identities = 153/192 (79%), Positives = 172/192 (88%), Gaps = 2/192 (1%)
Frame = +3
Query: 395 AISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGA 454
AISY++M +K+PEYRNGKL+IPCNSLSHALGIFQ GKF RD+LKLEAQAVTS++ EEGA
Sbjct: 6 AISYYLMNSKDPEYRNGKLVIPCNSLSHALGIFQEGKFSVRDTLKLEAQAVTSQR-EEGA 182
Query: 455 AVKPEGKTESPAPAVKTEAEIP-TSVRKTDGENSVPASKAEVP-DNEFEKRIRPEVIPAN 512
V+PE K E+PA +K E++ TSV KTDGEN+VP SK EVP DNEFEKRIRPEVIPAN
Sbjct: 183 LVEPEKKAENPASDIKAESDTSSTSVVKTDGENAVPESKVEVPPDNEFEKRIRPEVIPAN 362
Query: 513 EIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKA 572
EIGV FSD+GALDETK+SLQELVMLPLRRPDLF GGLLKPC+GILLFGPPGTGKTMLAKA
Sbjct: 363 EIGVKFSDVGALDETKESLQELVMLPLRRPDLFRGGLLKPCKGILLFGPPGTGKTMLAKA 542
Query: 573 IANEAGASFINV 584
IA+E+GASFINV
Sbjct: 543 IASESGASFINV 578
>TC229555 similar to GB|AAM62497.1|21553404|AY085265 26S proteasome
regulatory particle chain RPT6-like protein {Arabidopsis
thaliana;} , partial (85%)
Length = 1155
Score = 258 bits (660), Expect = 6e-69
Identities = 146/310 (47%), Positives = 197/310 (63%), Gaps = 6/310 (1%)
Frame = +2
Query: 495 VPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGG-LLKPC 553
+ N +E I ++I + I V F+ IG L+ K +L ELV+LPL+RPDLF G LL P
Sbjct: 170 IQTNPYEDVIACDIINPDHIDVEFNSIGGLETIKQALFELVILPLKRPDLFSHGKLLGPQ 349
Query: 554 RGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSP 613
+G+LL+GPPGTGKTMLAKAIA E+ A FINV +S + SKWFG+ +K V A+F+LA K+ P
Sbjct: 350 KGVLLYGPPGTGKTMLAKAIAKESRAVFINVRISNLMSKWFGDAQKLVAAVFSLAYKLQP 529
Query: 614 TIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDE 673
IIF+DEVDS LG+R R +HEAM +K EFM+ WDG T+ +++VLAATNRP +LDE
Sbjct: 530 AIIFIDEVDSFLGKR-RGTDHEAMLNMKTEFMALWDGFTTDQNAQVMVLAATNRPSELDE 706
Query: 674 AIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTT 733
AI+RR + +G+P R IL+ +L E+V +DF +A + EGY+GSDL +LC
Sbjct: 707 AILRRLPQAFEIGIPDQRERAEILKVVLKGERVEDNIDFGHIAGLCEGYTGSDLFDLCKK 886
Query: 734 AAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMA-- 791
AAY P+REL+ +E KK K + RPL+ DF+ A
Sbjct: 887 AAYFPIRELLDEE-----KKGKQSHAP--------------------RPLSQLDFEKALA 991
Query: 792 ---KSQVAAS 798
K++VAAS
Sbjct: 992 TSKKTKVAAS 1021
>TC221009 homologue to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (18%)
Length = 696
Score = 251 bits (641), Expect = 9e-67
Identities = 127/142 (89%), Positives = 134/142 (93%)
Frame = +1
Query: 605 FTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAA 664
FTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM++WDGL +K +RILVLAA
Sbjct: 1 FTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLLTKQGERILVLAA 180
Query: 665 TNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSG 724
TNRPFDLDEAIIRRFERRIMVGLPS ENRE ILRTLLAKEKV L+FKE+ATMTEGY+G
Sbjct: 181 TNRPFDLDEAIIRRFERRIMVGLPSVENREKILRTLLAKEKVDNELEFKEIATMTEGYTG 360
Query: 725 SDLKNLCTTAAYRPVRELIQQE 746
SDLKNLCTTAAYRPVRELIQQE
Sbjct: 361 SDLKNLCTTAAYRPVRELIQQE 426
>BU578832
Length = 437
Score = 237 bits (605), Expect = 1e-62
Identities = 120/146 (82%), Positives = 136/146 (92%)
Frame = +1
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
EAGASFINVS+S ITSKWFGEDEKNVRALF+LAAKV+PTIIF+DEVDSMLG+RT+ GEHE
Sbjct: 1 EAGASFINVSISNITSKWFGEDEKNVRALFSLAAKVAPTIIFIDEVDSMLGKRTKYGEHE 180
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENREN 695
AMRKIKNEFM++WDG+ +K +RILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRE
Sbjct: 181 AMRKIKNEFMAHWDGILTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENREM 360
Query: 696 ILRTLLAKEKVHGGLDFKELATMTEG 721
IL+TLLAKEK + +DF EL+T+TEG
Sbjct: 361 ILKTLLAKEK-YEHIDFNELSTITEG 435
>CD487729 similar to PIR|T02901|T02 MSP1 protein homolog T13J8.110 -
Arabidopsis thaliana, partial (18%)
Length = 604
Score = 233 bits (594), Expect = 3e-61
Identities = 122/203 (60%), Positives = 144/203 (70%), Gaps = 1/203 (0%)
Frame = +1
Query: 308 NECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAAND 367
++C VDE LT LFPYNIEIK P+DE+ L WK Q E DMK IQ QDN+NHI EVLAAND
Sbjct: 10 DDC*EVDERLTVLFPYNIEIKAPKDETHLGCWKGQLEKDMKDIQFQDNRNHIAEVLAAND 189
Query: 368 LDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIF 427
+DC DL+SIC ADT++LSNYIEEI+VSA+SYH+M K+PEYRNGKL+I NSLSH L +F
Sbjct: 190 IDCDDLNSICHADTILLSNYIEEIVVSALSYHLMNTKDPEYRNGKLVISANSLSHGLSLF 369
Query: 428 QAGKFGDRDSLKLEAQAVTSEKK-EEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGEN 486
Q GK S L+ E E+ K E K ++ AP K+E E + K DGEN
Sbjct: 370 QEGK----SSGNLKTNESNKENSGEDITGAKNEMKCDNQAPENKSETEKSIPITKKDGEN 537
Query: 487 SVPASKAEVPDNEFEKRIRPEVI 509
+PA K EVPDNEFEKRIRPEVI
Sbjct: 538 PIPA-KVEVPDNEFEKRIRPEVI 603
>BM271487 similar to GP|15810167|gb At1g64110/F22C12_22 {Arabidopsis
thaliana}, partial (15%)
Length = 411
Score = 224 bits (570), Expect = 2e-58
Identities = 107/136 (78%), Positives = 122/136 (89%)
Frame = +2
Query: 272 DKLLCRSQRIYKLFQTMLTKLSGPILIIGSRILDSGNECKRVDEMLTSLFPYNIEIKPPE 331
D+LL +SQRIY LFQ ML KLSGP+LI+GSR++DSGN+ + VDE + SLFPYNIEI+PPE
Sbjct: 2 DRLLYKSQRIYNLFQKMLKKLSGPVLILGSRVIDSGNDYEEVDEKINSLFPYNIEIRPPE 181
Query: 332 DESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLSNYIEEI 391
DES LVSWKSQ E D+K IQ+QDNKNHIMEVLAANDLDC DLDSICV+DTMVLSNYIEEI
Sbjct: 182 DESHLVSWKSQLEEDLKMIQVQDNKNHIMEVLAANDLDCDDLDSICVSDTMVLSNYIEEI 361
Query: 392 IVSAISYHIMKNKEPE 407
IVSAISYH+MKNK+ E
Sbjct: 362 IVSAISYHLMKNKDTE 409
>TC219888 similar to UP|Q944N4 (Q944N4) Tobacco mosaic virus helicase
domain-binding protein (Fragment), partial (50%)
Length = 1119
Score = 211 bits (536), Expect = 1e-54
Identities = 121/299 (40%), Positives = 179/299 (59%), Gaps = 6/299 (2%)
Frame = +3
Query: 525 DETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINV 584
++ K +L E+V+LP +R DLF G L +P RG+LLFGPPG GKTMLAKA+A+E+ A+F NV
Sbjct: 3 EKAKQALMEMVILPTKRRDLFTG-LRRPARGLLLFGPPGNGKTMLAKAVASESQATFFNV 179
Query: 585 SMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEF 644
+ +++TSKW GE EK VR LF +A P++IF+DE+DS++ R E++A R++K+EF
Sbjct: 180 TAASLTSKWVGEAEKLVRTLFMVAISRQPSVIFIDEIDSIMSTRL-ANENDASRRLKSEF 356
Query: 645 MSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKE 704
+ +DG+TS +D ++V+ ATN+P +LD+A++RR +RI + LP EN+ R LL K
Sbjct: 357 LIQFDGVTSNPDDIVIVIGATNKPQELDDAVLRRLVKRIYIPLPD----ENV-RKLLLKH 521
Query: 705 KVHG------GLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAE 758
K+ G D + L TEGYSGSDL+ LC AA P+REL
Sbjct: 522 KLKGQAFSLPSRDLERLVKETEGYSGSDLQALCEEAAMMPIREL---------------- 653
Query: 759 GQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYG 817
++ + +R L +DFK A + + S + EL +WN+ +G
Sbjct: 654 ----------GADILTVKANQVRGLRYEDFKKAMATIRPSL--NKSKWEELERWNEEFG 794
>TC208420 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (79%)
Length = 1523
Score = 202 bits (514), Expect = 5e-52
Identities = 116/282 (41%), Positives = 169/282 (59%), Gaps = 5/282 (1%)
Frame = +1
Query: 469 VKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEI----GVTFSDIGAL 524
+ T +PT V + ++ +P P E R E + + I V + I L
Sbjct: 319 IHTNGFVPTIVDERPQKSLLP------PFESAEMRALAESLSRDIIRGSPDVKWESIKGL 480
Query: 525 DETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINV 584
+ K L+E V++P++ P F G LL P +GILLFGPPGTGKTMLAKA+A E +F N+
Sbjct: 481 ENAKRLLKEAVVMPIKYPKYFTG-LLSPWKGILLFGPPGTGKTMLAKAVATECKTTFFNI 657
Query: 585 SMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRV-GEHEAMRKIKNE 643
S S++ SKW G+ EK V+ LF LA +P+ IF+DE+D+++ QR EHEA R++K E
Sbjct: 658 SASSVVSKWRGDSEKLVKVLFELARHHAPSTIFLDEIDAIISQRGEARSEHEASRRLKTE 837
Query: 644 FMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAK 703
+ DGLT K+++ + VLAATN P++LD A++RR E+RI+V LP R + LL +
Sbjct: 838 LLIQMDGLT-KTDELVFVLAATNLPWELDAAMLRRLEKRILVPLPEPVARRAMFEELLPQ 1014
Query: 704 EKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQ 745
+ + + L TEGYSGSD++ LC A +P+R L+ Q
Sbjct: 1015QPDEEPIPYDILVDKTEGYSGSDIRLLCKETAMQPLRRLMSQ 1140
>TC207657 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic reticulum
ATPase-like, partial (37%)
Length = 919
Score = 199 bits (506), Expect = 4e-51
Identities = 101/197 (51%), Positives = 127/197 (64%)
Frame = +3
Query: 633 EHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAEN 692
EHEA R+++NEFM+ WDGL SK RIL+L ATNRPFDLD+A+IRR RRI V LP AEN
Sbjct: 27 EHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAEN 206
Query: 693 RENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSK 752
R ILR LA+E ++ F +LA +T+GYSGSDLKNLC AAYRPV+EL+++E
Sbjct: 207 RMKILRIFLAQENLNSDFQFDKLANLTDGYSGSDLKNLCIAAAYRPVQELLEEE------ 368
Query: 753 KKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQW 812
K+ + LRPLN+ DF AKS+V S A + MNELR+W
Sbjct: 369 ----------------KKGASNDTTSILRPLNLDDFIQAKSKVGPSVAYDATSMNELRKW 500
Query: 813 NDLYGEGGSRKKEQLSY 829
N++YGEGGSR K +
Sbjct: 501 NEMYGEGGSRTKAPFGF 551
>TC204689 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase,
partial (98%)
Length = 1726
Score = 192 bits (487), Expect = 7e-49
Identities = 115/295 (38%), Positives = 171/295 (56%), Gaps = 2/295 (0%)
Frame = +3
Query: 449 KKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEV 508
+ EE AV +G P PA +A + KT ++ E P+ +
Sbjct: 375 RAEEIRAVLDDG---GPGPASNGDAAVAARP-KTKPKDGEGGGDGEDPEQAKLRAGLNSA 542
Query: 509 IPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTM 568
I + V ++D+ L+ K +LQE V+LP++ P F G +P R LL+GPPGTGK+
Sbjct: 543 IIREKPNVKWNDVAGLESAKQALQEAVILPVKFPQFFTGKR-RPWRAFLLYGPPGTGKSY 719
Query: 569 LAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR 628
LAKA+A EA ++F +VS S + SKW GE EK V LF +A + +P+IIF+DE+DS+ GQR
Sbjct: 720 LAKAVATEAESTFFSVSSSDLVSKWMGESEKLVSNLFEMARESAPSIIFIDEIDSLCGQR 899
Query: 629 TRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLP 688
E EA R+IK E + G+ ++ ++LVLAATN P+ LD+AI RRF++RI + LP
Sbjct: 900 GEGNESEASRRIKTELLVQMQGV-GHNDQKVLVLAATNTPYALDQAIRRRFDKRIYIPLP 1076
Query: 689 SAENRENILRTLLAKEKVHG--GLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE 741
+ R+++ + L + H DF+ LA+ TEG+SGSD+ + PVR+
Sbjct: 1077DLKARQHMFKVHLG-DTPHNLTESDFEYLASRTEGFSGSDISVCVKDVLFEPVRK 1238
>TC204690 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase,
partial (94%)
Length = 1630
Score = 192 bits (487), Expect = 7e-49
Identities = 114/286 (39%), Positives = 163/286 (56%), Gaps = 2/286 (0%)
Frame = +2
Query: 458 PEGKTESPAPAVKTEAEIPTSVRKTDGENSVPA-SKAEVPDNEFEKRIRPEVIPANEIGV 516
P G S AV T + DG+ P +K N R +P V
Sbjct: 362 PSGPASSGDAAVATRPKTKPKDGGKDGDGEDPEQAKLRAGLNSAIVREKPNV-------- 517
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANE 576
++D+ L+ K +LQE V+LP++ P F G +P R LL+GPPGTGK+ LAKA+A E
Sbjct: 518 KWNDVAGLESAKQALQEAVILPVKFPQFFTGKR-RPWRAFLLYGPPGTGKSYLAKAVATE 694
Query: 577 AGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEA 636
A ++F +VS S + SKW GE EK V LF +A + +P+IIFVDE+DS+ GQR E EA
Sbjct: 695 ADSTFFSVSSSDLVSKWMGESEKLVSNLFQMARESAPSIIFVDEIDSLCGQRGEGNESEA 874
Query: 637 MRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENI 696
R+IK E + G+ ++ ++LVLAATN P+ LD+AI RRF++RI + LP + R+++
Sbjct: 875 SRRIKTELLVQMQGV-GHNDQKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDLKARQHM 1051
Query: 697 LRTLLAKEKVH-GGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE 741
+ L + DF+ LA TEG+SGSD+ + PVR+
Sbjct: 1052FKVHLGDTPHNLAESDFEHLARKTEGFSGSDISVCVKDVLFEPVRK 1189
>TC206116 UP|CC48_SOYBN (P54774) Cell division cycle protein 48 homolog
(Valosin containing protein homolog) (VCP), complete
Length = 2583
Score = 186 bits (471), Expect = 5e-47
Identities = 101/297 (34%), Positives = 170/297 (57%), Gaps = 7/297 (2%)
Frame = +1
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V++ DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 1453 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIAN 1632
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR-TRVGE- 633
E A+FI+V + + WFGE E NVR +F A + +P ++F DE+DS+ QR + VG+
Sbjct: 1633 ECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGSSVGDA 1812
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
A ++ N+ ++ DG+++K + ++ ATNRP +D A++R R ++ I + LP +
Sbjct: 1813 GGAADRVLNQLLTEMDGMSAKK--TVFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDED 1986
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
+R I + L K + +D + LA T+G+SG+D+ +C A +RE I+++I+++
Sbjct: 1987 SRHQIFKACLRKSPIAKNVDLRALARHTQGFSGADITEICQRACKYAIRENIEKDIERER 2166
Query: 752 KKKKDAEGQNSQDAQDAKEEVEQ---ERVITLRPLNMQDFKMAKSQVAASFAAEGAG 805
K +++ E + D E++ E + ++ D + K Q A + G
Sbjct: 2167 KSRENPEAMDEDTVDDEVAEIKAAHFEESMKFARRSVSDADIRKYQAFAQTLQQSRG 2337
Score = 174 bits (441), Expect = 1e-43
Identities = 111/286 (38%), Positives = 165/286 (56%), Gaps = 2/286 (0%)
Frame = +1
Query: 479 VRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLP 538
V GE V A E+ E E R + +E+G + D+G + + ++ELV LP
Sbjct: 532 VETDPGEYCVVAPDTEI-FCEGEPLKREDEERLDEVG--YDDVGGVRKQMAQIRELVELP 702
Query: 539 LRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDE 598
LR P LF+ +KP +GILL+GPPG+GKT++A+A+ANE GA F ++ I SK GE E
Sbjct: 703 LRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCINGPEIMSKLAGESE 882
Query: 599 KNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDR 658
N+R F A K +P+IIF+DE+DS+ +R + E R+I ++ ++ DGL KS
Sbjct: 883 SNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HGEVERRIVSQLLTLMDGL--KSRAH 1053
Query: 659 ILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELA 716
++V+ ATNRP +D A+ R RF+R I +G+P R +LR K+ +D + +A
Sbjct: 1054VIVIGATNRPNSIDPALRRFGRFDREIDIGVPDEVGRLEVLRIHTKNMKLSDDVDLERIA 1233
Query: 717 TMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNS 762
T GY G+DL LCT AA + +RE + ++ + DAE NS
Sbjct: 1234KDTHGYVGADLAALCTEAALQCIRE--KMDVIDLEDETIDAEVLNS 1365
>TC230208 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic reticulum
ATPase-like, partial (45%)
Length = 642
Score = 170 bits (431), Expect = 2e-42
Identities = 81/129 (62%), Positives = 106/129 (81%), Gaps = 1/129 (0%)
Frame = +1
Query: 498 NEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFE-GGLLKPCRGI 556
+EFE V+P EIGV F DIGAL++ K +L ELV+LP+RRP+LF G LL+PC+GI
Sbjct: 256 DEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFSRGNLLRPCKGI 435
Query: 557 LLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTII 616
LLFGPPGTGKT+LAKA+A EAGA+FI+++ ST+TSKWFG+ EK +ALF+ A+K++P I+
Sbjct: 436 LLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSFASKLAPVIV 615
Query: 617 FVDEVDSML 625
FVDEVDS+L
Sbjct: 616 FVDEVDSLL 642
Score = 29.6 bits (65), Expect = 5.7
Identities = 26/89 (29%), Positives = 38/89 (42%)
Frame = +1
Query: 35 SSSSNALTPDKIENEMLRLVVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSK 94
S S L D+ E+ + VV E V FD+ E + L + ++ E+
Sbjct: 229 SQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGAL--EDVKKALNELVILPMRRPELFS 402
Query: 95 YTRNLAPASRTILLSGPAELYQQVLAKAL 123
L P + ILL GP + +LAKAL
Sbjct: 403 RGNLLRPC-KGILLFGPPGTGKTLLAKAL 486
>TC225683 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit 4-like
protein (26S proteasome subunit AtRPT2a), partial (97%)
Length = 1513
Score = 155 bits (391), Expect = 9e-38
Identities = 89/245 (36%), Positives = 143/245 (58%), Gaps = 5/245 (2%)
Frame = +2
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANE 576
+++DIG LD ++E V LPL P+L+E +KP +G++L+G PGTGKT+LAKA+AN
Sbjct: 566 SYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANS 745
Query: 577 AGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRV---GE 633
A+F+ V S + K+ G+ K VR LF +A +SP+I+F+DE+D++ +R GE
Sbjct: 746 TSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAHSGGE 925
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
E R + E ++ DG S+ + + V+ ATNR LD A++R R +R+I LP +
Sbjct: 926 REIQRTML-ELLNQLDGFDSRGD--VKVILATNRIESLDPALLRPGRIDRKIEFPLPDIK 1096
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
R I + ++ + ++ +E + +SG+D+K +CT A +RE + D
Sbjct: 1097TRRRIFQIHTSRMTLADDVNLEEFVMTKDEFSGADIKAICTEAGLLALRERRMKVTHADF 1276
Query: 752 KKKKD 756
KK KD
Sbjct: 1277KKAKD 1291
>TC225681 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit 4-like
protein (26S proteasome subunit AtRPT2a), partial (93%)
Length = 1433
Score = 155 bits (391), Expect = 9e-38
Identities = 89/245 (36%), Positives = 143/245 (58%), Gaps = 5/245 (2%)
Frame = +1
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANE 576
+++DIG LD ++E V LPL P+L+E +KP +G++L+G PGTGKT+LAKA+AN
Sbjct: 463 SYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANS 642
Query: 577 AGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRV---GE 633
A+F+ V S + K+ G+ K VR LF +A +SP+I+F+DE+D++ +R GE
Sbjct: 643 TSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAHSGGE 822
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
E R + E ++ DG S+ + + V+ ATNR LD A++R R +R+I LP +
Sbjct: 823 REIQRTML-ELLNQLDGFDSRGD--VKVILATNRIESLDPALLRPGRIDRKIEFPLPDIK 993
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
R I + ++ + ++ +E + +SG+D+K +CT A +RE + D
Sbjct: 994 TRRRIFQIHTSRMTLADDVNLEEFVMTKDEFSGADIKAICTEAGLLALRERRMKVTHADF 1173
Query: 752 KKKKD 756
KK KD
Sbjct: 1174KKAKD 1188
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,514,502
Number of Sequences: 63676
Number of extensions: 326811
Number of successful extensions: 1651
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 1508
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1563
length of query: 831
length of database: 12,639,632
effective HSP length: 105
effective length of query: 726
effective length of database: 5,953,652
effective search space: 4322351352
effective search space used: 4322351352
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140913.6


