

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.5 + phase: 0 /pseudo
(584 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
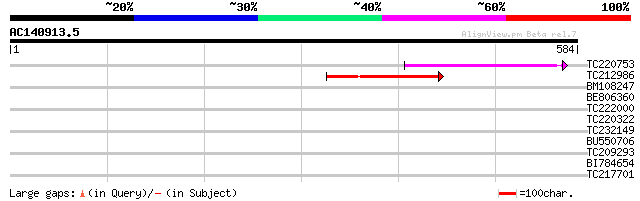
Score E
Sequences producing significant alignments: (bits) Value
TC220753 104 1e-22
TC212986 103 2e-22
BM108247 weakly similar to PIR|H84710|H847 Mutator-like transpos... 40 0.003
BE806360 similar to GP|6175165|gb| Mutator-like transposase {Ara... 35 0.071
TC222000 weakly similar to PIR|H84710|H84710 Mutator-like transp... 35 0.12
TC220322 33 0.27
TC232149 33 0.46
BU550706 weakly similar to GP|23093901|gb CG32356-PA {Drosophila... 30 3.9
TC209293 homologue to UP|RN12_HUMAN (Q9NVW2) RING finger protein... 29 6.7
BI784654 GP|27355006| AcrB/AcrD/AcrF family protein {Bradyrhizob... 28 8.7
TC217701 homologue to GB|AAL06971.1|15810091|AY056083 AT4g30260/... 28 8.7
>TC220753
Length = 1312
Score = 104 bits (260), Expect = 1e-22
Identities = 58/168 (34%), Positives = 88/168 (51%)
Frame = +2
Query: 407 VGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREIVNYREGRPMRTFKVDLRA 466
+ +G ++E ++++ + +H+V + RE V+EI N R GR V L
Sbjct: 50 IKAGHRYSEDVYVMMQENQHIATSHYVRMYVRETKEXEVQEIANTRLGRRGMACTVKLNE 229
Query: 467 GWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNESVYSIYNTTFKVVQDKSYWL 526
CD G+FQAL LPCSH IAAC+ +Y + V K E+++ +Y F + + W
Sbjct: 230 WLCDYGQFQALRLPCSHAIAACAFCNLNYDDFVDPVYKLENIFKVYQHHFHSLGSEDTWP 409
Query: 527 PYDGPVLCHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPIPRQCWLCR 574
Y GP +P+ RR GRP +TRI EMDE + R ++C LC+
Sbjct: 410 QYLGPHFMSDPSKRRQISGRPATTRIHNEMDESIPNRL---KKCSLCK 544
>TC212986
Length = 404
Score = 103 bits (257), Expect = 2e-22
Identities = 48/121 (39%), Positives = 77/121 (62%)
Frame = +1
Query: 327 GVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQ 386
G I+ A +W+D +PK+ W Q +DEG+RWG +T+N+ ES N +FK TR+L V+ +V+
Sbjct: 16 GDIRANKPSAAEWLDQLPKQKWVQCFDEGKRWG-LTTNLSESVNSMFKNTRHLSVSSLVE 192
Query: 387 STYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVR 446
TY++ A FA+R ++ + SG ++E +AI +SNTH V +F+R +TF +
Sbjct: 193 ETYFKTAQLFANRGRQTQAMINSGSQYSEVVFDAINSGQQESNTHIVNEFDRHNHTFIIT 372
Query: 447 E 447
E
Sbjct: 373 E 375
>BM108247 weakly similar to PIR|H84710|H847 Mutator-like transposase
[imported] - Arabidopsis thaliana, partial (4%)
Length = 265
Score = 40.0 bits (92), Expect = 0.003
Identities = 23/81 (28%), Positives = 39/81 (47%), Gaps = 4/81 (4%)
Frame = +2
Query: 309 KLVQLIY----ALNVPLFDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSN 364
+LV L++ A F G I++ + A W+ W + +G R+GH++SN
Sbjct: 11 RLVHLLWKASHATTTIAFKEKMGEIEEVSPEAAKWLQQFXPSQWALVHFKGTRFGHLSSN 190
Query: 365 IVESWNFVFKGTRNLPVTPIV 385
I E +N TR LP+ ++
Sbjct: 191 I-EEFNKWILDTRELPIIXVI 250
>BE806360 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (17%)
Length = 431
Score = 35.4 bits (80), Expect = 0.071
Identities = 14/21 (66%), Positives = 17/21 (80%)
Frame = +2
Query: 348 WTQAYDEGRRWGHMTSNIVES 368
W AY EG R+GH+T+NIVES
Sbjct: 350 WATAYFEGHRFGHLTANIVES 412
>TC222000 weakly similar to PIR|H84710|H84710 Mutator-like transposase
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (12%)
Length = 617
Score = 34.7 bits (78), Expect = 0.12
Identities = 26/103 (25%), Positives = 36/103 (34%), Gaps = 5/103 (4%)
Frame = +2
Query: 462 VDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNESVYSIYNTTFKVVQD 521
V++ + C C +Q +PCSH AA S R D S Y T +
Sbjct: 20 VNIGSHSCSCRDWQLNGIPCSHAAAALISCRKDVYAFSQKCFTAASFRDTYAETIHPIPG 199
Query: 522 KSYW-----LPYDGPVLCHNPNMRRLKKGRPNSTRIRTEMDEE 559
K W D +L P R GRP +++ E
Sbjct: 200 KLEWSKTGNSSMDDNILVVRPPKLRRPPGRPEKRMCVDDLNRE 328
>TC220322
Length = 1264
Score = 33.5 bits (75), Expect = 0.27
Identities = 40/178 (22%), Positives = 74/178 (41%), Gaps = 3/178 (1%)
Frame = +2
Query: 312 QLIYALNVPLFDYYRGVIKKANQRALDWVDNIPKENWTQA-YDEGRRWGHMTSNIVESWN 370
Q + V F+ + +K N +A ++++ PK+ WT+A + + ++ +N E +N
Sbjct: 2 QCAKSTTVAEFEGHMAHLKTINCQAWEYLNKWPKQAWTKAHFSTIPKVDNICNNTCEVFN 181
Query: 371 FVFKGTRNLPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNT 430
F R P+ +++ + A R K + G L ++ ++
Sbjct: 182 FRILQYRCKPIITMLEEIRSYIMRTMAARKVKLSGKPGPLCL--------VQYKRLEKEF 337
Query: 431 HHVEQFNRERYTFSVREIVNYREGRPMRTFKVDLR-AGW-CDCGKFQALHLPCSHVIA 486
H Q+ + + R M KV++ AGW C CG +Q +PC H IA
Sbjct: 338 HFANQWT----PIWCGDNMGLRYEVHMWGNKVEVNLAGWTCTCGVWQLTGMPCRHAIA 499
>TC232149
Length = 1005
Score = 32.7 bits (73), Expect = 0.46
Identities = 12/31 (38%), Positives = 22/31 (70%)
Frame = -1
Query: 322 FDYYRGVIKKANQRALDWVDNIPKENWTQAY 352
F+ V+K+ N++A ++D P+E+WT+AY
Sbjct: 585 FNVIMKVVKRMNEKAWAYLDKGPRESWTKAY 493
>BU550706 weakly similar to GP|23093901|gb CG32356-PA {Drosophila
melanogaster}, partial (1%)
Length = 572
Score = 29.6 bits (65), Expect = 3.9
Identities = 25/76 (32%), Positives = 35/76 (45%)
Frame = +1
Query: 357 RWGHMTSNIVESWNFVFKGTRNLPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEY 416
RW +S + SW V + R V P+ + +C + A K VRVGSG T +
Sbjct: 262 RW---SSPSLPSWCRVLRSVR---VVPLEFACCCSCSCCSSPFACKNRVRVGSGQPNTTH 423
Query: 417 CQNAIKDDIVKSNTHH 432
C+N K K+ HH
Sbjct: 424 CENFKKTK--KAVFHH 465
>TC209293 homologue to UP|RN12_HUMAN (Q9NVW2) RING finger protein 12 (LIM
domain interacting RING finger protein) (RING finger LIM
domain-binding protein) (R-LIM) (NY-REN-43 antigen),
partial (4%)
Length = 957
Score = 28.9 bits (63), Expect = 6.7
Identities = 22/73 (30%), Positives = 32/73 (43%), Gaps = 16/73 (21%)
Frame = -2
Query: 526 LPYDG--PVLCHNPNMRRLKKGRPNSTRIRTEMDEEVV--------------ERTPIPRQ 569
LP G P+L NP + + K +P TR T + + + R P P +
Sbjct: 755 LPTTGAAPILLENPLVAKEAKTKPTETRHETRVRQIEIFTANWARGWGGGASPRHPTPIR 576
Query: 570 CWLCRHTVILREI 582
W HTV+LR+I
Sbjct: 575 VW---HTVLLRQI 546
>BI784654 GP|27355006| AcrB/AcrD/AcrF family protein {Bradyrhizobium
japonicum USDA 110}, partial (13%)
Length = 421
Score = 28.5 bits (62), Expect = 8.7
Identities = 15/38 (39%), Positives = 17/38 (44%)
Frame = +3
Query: 537 PNMRRLKKGRPNSTRIRTEMDEEVVERTPIPRQCWLCR 574
P R + GRP S R R P PR+ WLCR
Sbjct: 69 PASRPARAGRPRSASCRP--------RAPRPRRIWLCR 158
>TC217701 homologue to GB|AAL06971.1|15810091|AY056083 AT4g30260/F9N11_110
{Arabidopsis thaliana;} , partial (68%)
Length = 878
Score = 28.5 bits (62), Expect = 8.7
Identities = 11/34 (32%), Positives = 18/34 (52%)
Frame = -1
Query: 534 CHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPIP 567
C++ N+R +GRP + +EE + PIP
Sbjct: 365 CNSKNLRLFDRGRPREGEAEEDNEEEWAPQIPIP 264
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.339 0.146 0.487
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,056,104
Number of Sequences: 63676
Number of extensions: 353685
Number of successful extensions: 2699
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 2684
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2698
length of query: 584
length of database: 12,639,632
effective HSP length: 102
effective length of query: 482
effective length of database: 6,144,680
effective search space: 2961735760
effective search space used: 2961735760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140913.5


