

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.4 - phase: 0
(374 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
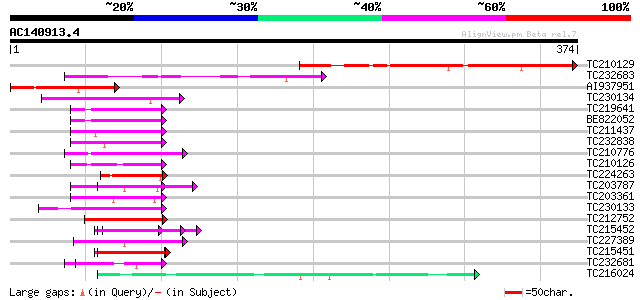
Score E
Sequences producing significant alignments: (bits) Value
TC210129 similar to UP|Q9SH58 (Q9SH58) F22C12.16, partial (17%) 191 5e-49
TC232683 similar to UP|Q9SH58 (Q9SH58) F22C12.16, partial (5%) 91 6e-19
AI937951 81 7e-16
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 52 4e-07
TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 49 3e-06
BE822052 similar to GP|4557063|gb|A expressed protein {Arabidops... 49 4e-06
TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 49 5e-06
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 47 1e-05
TC210776 similar to UP|Q8H6B1 (Q8H6B1) Global transcription fact... 46 3e-05
TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C2... 46 3e-05
TC224263 weakly similar to UP|Q6GQH4 (Q6GQH4) Egr1 protein, part... 46 3e-05
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 45 4e-05
TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 45 4e-05
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 45 5e-05
TC212752 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific... 45 7e-05
TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%) 44 9e-05
TC227389 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacety... 44 1e-04
TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembl... 44 2e-04
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 44 2e-04
TC216024 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F2... 43 2e-04
>TC210129 similar to UP|Q9SH58 (Q9SH58) F22C12.16, partial (17%)
Length = 919
Score = 191 bits (485), Expect = 5e-49
Identities = 119/188 (63%), Positives = 128/188 (67%), Gaps = 5/188 (2%)
Frame = +1
Query: 192 KQHKPERIKFKVEEVPIVSLFTRDNSSKNSNKSQTNQNQKVEHESSLSPLEEKRFSKEIV 251
K K +KFKVEEVPIVSLF S+K+ NQNQK E P EEKRFSKE V
Sbjct: 10 KDKKHFTVKFKVEEVPIVSLF--------SSKANKNQNQKQTSEDK--PSEEKRFSKE-V 156
Query: 252 MHKYLKMVKPLYIRVSRRYSHGDKSSPSEKVEAETAT---EGETVAENEENNAKTQSQKQ 308
MHKYLKMVKPLYIRVSRRY +P+ K T EG +V E N +S Q
Sbjct: 157 MHKYLKMVKPLYIRVSRRYGEKLSLNPAAKAPPPPPTAEPEGVSVGGGETN---VKSSTQ 327
Query: 309 GNNIPAGLRVVYKHLGKSRSASSAVAAS--PPVLVSSKRRDDSLLQQQDGIQGAILHCKR 366
G +PAGL VV KHLGKSRSASSAVAA+ P V VSSKRRDDSLLQQQDGIQGAILHCKR
Sbjct: 328 GQKLPAGLSVVCKHLGKSRSASSAVAAAAPPAVFVSSKRRDDSLLQQQDGIQGAILHCKR 507
Query: 367 SFNASRGE 374
SFNASR E
Sbjct: 508 SFNASRPE 531
>TC232683 similar to UP|Q9SH58 (Q9SH58) F22C12.16, partial (5%)
Length = 420
Score = 91.3 bits (225), Expect = 6e-19
Identities = 65/175 (37%), Positives = 83/175 (47%), Gaps = 2/175 (1%)
Frame = +1
Query: 37 LTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
L + + D D +GPFFD+E P+ EED D++D+++ D
Sbjct: 61 LASPDIDADTDADGPFFDVELAVPDSEED----------------------DTNDNNNSD 174
Query: 97 SGETESEFKITLSPSNNERSDANLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEP 156
S +E EFK+TLSPS + S+ FI N+ NNS
Sbjct: 175 SA-SEKEFKLTLSPSTS-------------------------SNDFIFNSQPNNS----- 261
Query: 157 NNKPQFTASLLKSATKFRVFMSGLK--KPKTDSVLEKKQHKPERIKFKVEEVPIV 209
KPQFTASL KSATK RVFM GLK KP + K+ K +KFK EE PIV
Sbjct: 262 --KPQFTASLFKSATKLRVFMLGLKKLKPNAPAAPNLKKKKLFTVKFKAEEYPIV 420
>AI937951
Length = 418
Score = 81.3 bits (199), Expect = 7e-16
Identities = 41/76 (53%), Positives = 52/76 (67%), Gaps = 4/76 (5%)
Frame = +3
Query: 1 MEAFSLLKYWRGGGAVVGLSPSSDSTTTTNVNTTTILTTAESSD----DNDDEGPFFDLE 56
MEA +LLKYWRGGGA+ G PSSD+ ++ TTTILT E+ + D+DD+GPFFDLE
Sbjct: 186 MEAXTLLKYWRGGGAL-GHPPSSDTPSSRTATTTTILTAVENENATESDDDDDGPFFDLE 362
Query: 57 FTAPEEEEDGFEETNH 72
FT P+E+ NH
Sbjct: 363 FTVPDEDAHENPNANH 410
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 52.0 bits (123), Expect = 4e-07
Identities = 27/98 (27%), Positives = 50/98 (50%), Gaps = 4/98 (4%)
Frame = +2
Query: 22 SSDSTTTTNVNTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEK 81
+ D++ T + + + E DD+D++ F + E + D E N + ++++
Sbjct: 260 NKDASDTEDDDDDDDVNDGEDGDDDDEDDEDFSGDDGGEEADSDDDPEANGGGESDDDDE 439
Query: 82 DEDDDEDSDD----DDDDDSGETESEFKITLSPSNNER 115
D+DDD+D +D D+DDD E E E + T P + +R
Sbjct: 440 DDDDDDDDNDEDDGDEDDDDEEDEDEDEETPQPPSKKR 553
Score = 37.4 bits (85), Expect = 0.011
Identities = 20/70 (28%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Frame = +2
Query: 36 ILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDED-SDDDDD 94
++T + +D N+ P D F P ++ + T+ +N+ + +D+DDD+D +D +D
Sbjct: 152 VVTGSLLTDANNLVSPMVDGRF--PLDQPCKGKHTSEENKDASDTEDDDDDDDVNDGEDG 325
Query: 95 DDSGETESEF 104
DD E + +F
Sbjct: 326 DDDDEDDEDF 355
Score = 30.4 bits (67), Expect = 1.4
Identities = 23/83 (27%), Positives = 37/83 (43%)
Frame = -3
Query: 89 SDDDDDDDSGETESEFKITLSPSNNERSDANLSLSPSEDLFFKGKLVQLKSSSFILNNNN 148
S S + S +LS S++ S ++ SLSP G + SS +
Sbjct: 522 SSSSSSSSSSSSSSPSSSSLSSSSSSSSSSSSSLSPPP--LASGSSSESASSPPSSPEKS 349
Query: 149 NNSSSSEPNNKPQFTASLLKSAT 171
++SSSS + P FT+S S++
Sbjct: 348 SSSSSSSSPSSPSFTSSSSSSSS 280
>TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 942
Score = 49.3 bits (116), Expect = 3e-06
Identities = 24/63 (38%), Positives = 36/63 (57%)
Frame = +1
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGET 100
++ D +D+EG D +F+ EEE E+ N + D DDD+D DDDDDD+G+
Sbjct: 421 DADDQDDEEG---DEDFSGEGEEEADPEDDPEANGAGGSDSDGDDDDDGGDDDDDDNGDD 591
Query: 101 ESE 103
+ E
Sbjct: 592 DGE 600
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/77 (31%), Positives = 40/77 (51%), Gaps = 8/77 (10%)
Frame = +1
Query: 35 TILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDD-------- 86
T L ++SD ++D D + +++E+G E+ + + EEE D +DD
Sbjct: 355 TYLLNKDASDSDEDGDEEEDDDDADDQDDEEGDEDFSGEG---EEEADPEDDPEANGAGG 525
Query: 87 EDSDDDDDDDSGETESE 103
DSD DDDDD G+ + +
Sbjct: 526 SDSDGDDDDDGGDDDDD 576
Score = 30.0 bits (66), Expect = 1.8
Identities = 13/46 (28%), Positives = 21/46 (45%)
Frame = +1
Query: 72 HKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSD 117
H ++ D D+D D ++DDDD + + E S E +D
Sbjct: 352 HTYLLNKDASDSDEDGDEEEDDDDADDQDDEEGDEDFSGEGEEEAD 489
Score = 29.6 bits (65), Expect = 2.3
Identities = 12/49 (24%), Positives = 31/49 (62%)
Frame = +1
Query: 40 AESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDED 88
++S D+DD+G ++++D + + +++++ E++DE+D+ED
Sbjct: 526 SDSDGDDDDDG---------GDDDDDDNGDDDGEDEEEGEDEDEEDEED 645
>BE822052 similar to GP|4557063|gb|A expressed protein {Arabidopsis
thaliana}, partial (28%)
Length = 668
Score = 48.9 bits (115), Expect = 4e-06
Identities = 27/64 (42%), Positives = 36/64 (56%), Gaps = 1/64 (1%)
Frame = -1
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDD-DDSGE 99
+++D ND+EG D +F+ EEE E+ N D+D DED DDD+D DD GE
Sbjct: 485 DTNDQNDEEG---DEDFSGEGEEEADPEDDPEANGAGGSNDDDDSDEDDDDDNDGDDDGE 315
Query: 100 TESE 103
E E
Sbjct: 314 DEDE 303
Score = 35.0 bits (79), Expect = 0.055
Identities = 22/68 (32%), Positives = 38/68 (55%), Gaps = 4/68 (5%)
Frame = -1
Query: 40 AESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSD----DDDDD 95
A SD+++DE D T + +E+G E+ + + EEE D +DD +++ +DDD
Sbjct: 527 ASDSDEDEDEEDDHD---TNDQNDEEGDEDFSGEG---EEEADPEDDPEANGAGGSNDDD 366
Query: 96 DSGETESE 103
DS E + +
Sbjct: 365 DSDEDDDD 342
Score = 33.1 bits (74), Expect = 0.21
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Frame = -1
Query: 71 NHKNQQQEEEKDEDDDEDSDDDDDDD-----SGETESEFKITLSPSNNERSDANLSLSPS 125
N +E++DE+DD D++D +D++ SGE E E P N +N
Sbjct: 536 NKDASDSDEDEDEEDDHDTNDQNDEEGDEDFSGEGEEEADPEDDPEANGAGGSNDDDDSD 357
Query: 126 ED 127
ED
Sbjct: 356 ED 351
>TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 857
Score = 48.5 bits (114), Expect = 5e-06
Identities = 23/68 (33%), Positives = 38/68 (55%), Gaps = 5/68 (7%)
Frame = +1
Query: 41 ESSDDNDDEGPFFDL-----EFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDD 95
E D++D++GP D EF+ E +E G E + ++ ++DD +D+ D++DD
Sbjct: 379 EDGDEDDEDGPDEDEDGDDEEFSGEEGDEGGDPEDEANGEGSDDGDEDDDGDDNGDEEDD 558
Query: 96 DSGETESE 103
D GE E E
Sbjct: 559 DDGEDEEE 582
Score = 34.7 bits (78), Expect = 0.072
Identities = 16/45 (35%), Positives = 24/45 (52%)
Frame = +1
Query: 60 PEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEF 104
P E+ G + +N+ E E +ED DED +D D+D + EF
Sbjct: 310 PLEDLFGRKRAYLQNKNSETEDEEDGDEDDEDGPDEDEDGDDEEF 444
Score = 33.9 bits (76), Expect = 0.12
Identities = 20/57 (35%), Positives = 28/57 (49%)
Frame = +1
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSD 117
E+EEDG +E+ ++ DED D DD++ SGE E +N E SD
Sbjct: 370 EDEEDG-----------DEDDEDGPDEDEDGDDEEFSGEEGDEGGDPEDEANGEGSD 507
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 47.4 bits (111), Expect = 1e-05
Identities = 26/69 (37%), Positives = 40/69 (57%), Gaps = 6/69 (8%)
Frame = +2
Query: 41 ESSDDNDDEGPFFD-LEFTAPE-----EEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDD 94
E DD+DDEG D E AP+ ++ED EE + + + ++ D D D+DSDDD+D
Sbjct: 347 EEEDDDDDEGDDNDDEEDDAPDGGDDDDDEDDEEEGDVQRGGEPDDDDNDSDDDSDDDED 526
Query: 95 DDSGETESE 103
+D + E +
Sbjct: 527 EDEEDEEEQ 553
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/84 (27%), Positives = 44/84 (52%)
Frame = +2
Query: 44 DDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
+++D+E D E++ED EE + ++ + + +EDD D DDDDD+ E E +
Sbjct: 281 EEDDEEYESVDEVVDDAEDDEDE-EEDDDDDEGDDNDDEEDDAPDGGDDDDDEDDEEEGD 457
Query: 104 FKITLSPSNNERSDANLSLSPSED 127
+ P +++ +D++ ED
Sbjct: 458 VQRGGEPDDDD-NDSDDDSDDDED 526
Score = 37.0 bits (84), Expect = 0.014
Identities = 25/67 (37%), Positives = 33/67 (48%)
Frame = +2
Query: 37 LTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
L TAE + + D F+ E EEEED +E K + + K D D DSDDDD +
Sbjct: 596 LETAEEEEASSD----FEPEENGEEEEEDVDDEDCEKAEAPPKRKRSDKD-DSDDDDGGE 760
Query: 97 SGETESE 103
E S+
Sbjct: 761 DDERPSK 781
Score = 35.0 bits (79), Expect = 0.055
Identities = 15/64 (23%), Positives = 32/64 (49%)
Frame = +2
Query: 32 NTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDD 91
N +L + + + D E EEE+D +E + + ++++ D DD+D +D
Sbjct: 260 NEVKVLVEEDDEEYESVDEVVDDAEDDEDEEEDDDDDEGDDNDDEEDDAPDGGDDDDDED 439
Query: 92 DDDD 95
D+++
Sbjct: 440 DEEE 451
Score = 32.3 bits (72), Expect = 0.36
Identities = 20/77 (25%), Positives = 38/77 (48%)
Frame = +2
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGET 100
+ SDD++DE + E EEE+ G E + EEE+ D E ++ ++++
Sbjct: 503 DDSDDDEDEDE--EDEEEQGEEEDLGTEYLIRPLETAEEEEASSDFEPEENGEEEEEDVD 676
Query: 101 ESEFKITLSPSNNERSD 117
+ + + +P +RSD
Sbjct: 677 DEDCEKAEAPPKRKRSD 727
Score = 32.0 bits (71), Expect = 0.46
Identities = 27/95 (28%), Positives = 39/95 (40%), Gaps = 34/95 (35%)
Frame = +2
Query: 41 ESSDDNDD---------------EGPFFDL--EF------TAPEEEEDGFEETNHKNQQQ 77
+ +D+DD +G DL E+ TA EEE E +++
Sbjct: 482 DDDNDSDDDSDDDEDEDEEDEEEQGEEEDLGTEYLIRPLETAEEEEASSDFEPEENGEEE 661
Query: 78 EEEKDEDDDE-----------DSDDDDDDDSGETE 101
EE+ D++D E D DD DDDD GE +
Sbjct: 662 EEDVDDEDCEKAEAPPKRKRSDKDDSDDDDGGEDD 766
>TC210776 similar to UP|Q8H6B1 (Q8H6B1) Global transcription factor group C
102, partial (12%)
Length = 411
Score = 45.8 bits (107), Expect = 3e-05
Identities = 24/81 (29%), Positives = 42/81 (51%)
Frame = +1
Query: 37 LTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
L +S +N +E E + E E D +E + E E D+DDDEDS++D +++
Sbjct: 79 LEATDSESENSEESDK-GYEPSDVEPESDSEDEASDSESLVESEDDDDDDEDSEEDSEEE 255
Query: 97 SGETESEFKITLSPSNNERSD 117
G+T E + S ++ E+ +
Sbjct: 256 KGKTWEELEREASNADREKGN 318
Score = 29.6 bits (65), Expect = 2.3
Identities = 19/72 (26%), Positives = 33/72 (45%)
Frame = +1
Query: 19 LSPSSDSTTTTNVNTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQE 78
+ P SDS + + + L +E DD+D+ D E + EE+ +EE + +
Sbjct: 145 VEPESDSEDEASDSES--LVESEDDDDDDE-----DSEEDSEEEKGKTWEELEREASNAD 303
Query: 79 EEKDEDDDEDSD 90
EK + D + D
Sbjct: 304 REKGNESDSEED 339
>TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23), partial
(7%)
Length = 735
Score = 45.8 bits (107), Expect = 3e-05
Identities = 24/63 (38%), Positives = 36/63 (57%)
Frame = +3
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGET 100
E D DD+G D EF+ E +E+G E + + +DDD+D+D+++DDD GE
Sbjct: 372 EDGHDQDDDGE--DEEFSGEEGDEEGDPEDD---PEANGGGSDDDDDDNDNEEDDDDGED 536
Query: 101 ESE 103
E E
Sbjct: 537 EEE 545
Score = 37.4 bits (85), Expect = 0.011
Identities = 22/90 (24%), Positives = 47/90 (51%), Gaps = 6/90 (6%)
Frame = +3
Query: 43 SDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDS------DDDDDDD 96
++D+D +G D + ++++DG E+ ++ +EE D +DD ++ DDDDD+D
Sbjct: 336 TEDDDKDGDEDDED--GHDQDDDG-EDEEFSGEEGDEEGDPEDDPEANGGGSDDDDDDND 506
Query: 97 SGETESEFKITLSPSNNERSDANLSLSPSE 126
+ E + + + + E + S P++
Sbjct: 507 NEEDDDDGEDEEEEEDEEEDEEETSQPPTK 596
Score = 32.0 bits (71), Expect = 0.46
Identities = 12/36 (33%), Positives = 20/36 (55%)
Frame = +3
Query: 68 EETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
E N + + +++ DEDD++ D DDD + E E
Sbjct: 318 ENKNSETEDDDKDGDEDDEDGHDQDDDGEDEEFSGE 425
Score = 31.2 bits (69), Expect = 0.79
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 5/35 (14%)
Frame = +3
Query: 72 HKNQQQEEEKDEDDDEDSDDDDD-----DDSGETE 101
H + + + EDDD+D D+DD+ DD GE E
Sbjct: 306 HAYLENKNSETEDDDKDGDEDDEDGHDQDDDGEDE 410
Score = 30.8 bits (68), Expect = 1.0
Identities = 16/66 (24%), Positives = 36/66 (54%), Gaps = 3/66 (4%)
Frame = +3
Query: 42 SSDDNDDEG-PFFDLEFTAPEEEEDGFEETNHKNQQ--QEEEKDEDDDEDSDDDDDDDSG 98
S ++ D+EG P D E ++D + N ++ ++EE++ED++ED ++ +
Sbjct: 417 SGEEGDEEGDPEDDPEANGGGSDDDDDDNDNEEDDDDGEDEEEEEDEEEDEEETSQPPTK 596
Query: 99 ETESEF 104
+ + E+
Sbjct: 597 KRK*EY 614
Score = 29.3 bits (64), Expect = 3.0
Identities = 10/31 (32%), Positives = 18/31 (57%)
Frame = +3
Query: 73 KNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
K+ E + E +D+D D D+DD+ G + +
Sbjct: 303 KHAYLENKNSETEDDDKDGDEDDEDGHDQDD 395
>TC224263 weakly similar to UP|Q6GQH4 (Q6GQH4) Egr1 protein, partial (8%)
Length = 790
Score = 45.8 bits (107), Expect = 3e-05
Identities = 25/49 (51%), Positives = 30/49 (61%), Gaps = 5/49 (10%)
Frame = -2
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSG-----ETESEF 104
+E DG +E + E E DED+DEDSDDD D+DSG ETE EF
Sbjct: 657 KEAHDG-KEDEQGSDNDENEDDEDEDEDSDDDYDEDSGSEEYEETEEEF 514
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 45.4 bits (106), Expect = 4e-05
Identities = 21/66 (31%), Positives = 33/66 (49%)
Frame = -3
Query: 59 APEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSDA 118
APE G EE +++ ++ + +DDDED DDDD++D E + N E +
Sbjct: 490 APEGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEED 311
Query: 119 NLSLSP 124
+L P
Sbjct: 310 EEALQP 293
Score = 41.6 bits (96), Expect = 6e-04
Identities = 26/85 (30%), Positives = 41/85 (47%), Gaps = 22/85 (25%)
Frame = -3
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGF-EETNHKN-----------------QQQEEEKD 82
E DD+D +G F + E E+ G+ +N+K+ ++ E+E
Sbjct: 607 EGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKSNSKKAPEGGAGGAEENGEEDEDEDG 428
Query: 83 EDDDEDSDDDDDDD----SGETESE 103
+D D+D +DDDDDD +GE E E
Sbjct: 427 DDQDDDDEDDDDDDEEDEAGEEEDE 353
Score = 33.5 bits (75), Expect = 0.16
Identities = 18/47 (38%), Positives = 26/47 (55%)
Frame = -3
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDE 87
E DD DD+ D + EE+E G EE + +E E+DE+D+E
Sbjct: 436 EDGDDQDDDDEDDDDD---DEEDEAGEEEDEDGAEDEENEEDEEDEE 305
Score = 32.7 bits (73), Expect = 0.27
Identities = 16/62 (25%), Positives = 32/62 (50%), Gaps = 9/62 (14%)
Frame = -3
Query: 63 EEDGFEETNHKNQQQEEEKDEDDDED-------SDDDD--DDDSGETESEFKITLSPSNN 113
EE+G E+ + Q+++ ++DDD+D +D+D +D+ E + E + L P
Sbjct: 463 EENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEEDEEALQPPKK 284
Query: 114 ER 115
+
Sbjct: 283 RK 278
Score = 32.3 bits (72), Expect = 0.36
Identities = 12/58 (20%), Positives = 35/58 (59%)
Frame = -3
Query: 40 AESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDS 97
A +++N +E D + ++E+D ++ + ++E+E +D+E+ +D++D+++
Sbjct: 475 AGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEEDEEA 302
Score = 32.0 bits (71), Expect = 0.46
Identities = 20/72 (27%), Positives = 29/72 (39%), Gaps = 2/72 (2%)
Frame = -3
Query: 55 LEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSP--SN 112
L P + + ++ K + E E DD+ D DDD D GE E E N
Sbjct: 700 LLINTPTAKGESKIQSEDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSEDGGGYGN 521
Query: 113 NERSDANLSLSP 124
N + +N +P
Sbjct: 520 NSNNKSNSKKAP 485
>TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 1090
Score = 45.4 bits (106), Expect = 4e-05
Identities = 28/86 (32%), Positives = 38/86 (43%), Gaps = 23/86 (26%)
Frame = -1
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGF--------------------EETNHKNQQQEEE 80
E DD+D +G F + E E+ G+ E N + + E+
Sbjct: 574 EGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKSNSKKAPEGGAGGAEENGEEDEDEDG 395
Query: 81 KDEDDDEDSDDDDD---DDSGETESE 103
D+DDDED DDDDD D+ GE E E
Sbjct: 394 DDQDDDEDDDDDDDDEEDEGGEEEDE 317
Score = 35.8 bits (81), Expect = 0.032
Identities = 12/41 (29%), Positives = 27/41 (65%)
Frame = -1
Query: 63 EEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
EED E+ + ++ ++++ D+DD+ED +++D+ G + E
Sbjct: 418 EEDEDEDGDDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDEE 296
Score = 33.9 bits (76), Expect = 0.12
Identities = 13/50 (26%), Positives = 29/50 (58%)
Frame = -1
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSD 90
+ +D DD+ D + +EE++G EE + + EE +++++DE+ +
Sbjct: 412 DEDEDGDDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDEENEEDEEDEEEE 263
Score = 33.1 bits (74), Expect = 0.21
Identities = 12/56 (21%), Positives = 32/56 (56%)
Frame = -1
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
E DD DD+ ++++D +E + ++++E+ ED++ + D++D+++
Sbjct: 403 EDGDDQDDD----------EDDDDDDDDEEDEGGEEEDEDGAEDEENEEDEEDEEE 266
Score = 31.6 bits (70), Expect = 0.61
Identities = 18/58 (31%), Positives = 25/58 (43%), Gaps = 2/58 (3%)
Frame = -1
Query: 69 ETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSP--SNNERSDANLSLSP 124
++ K + E E DD+ D DDD D GE E E NN + +N +P
Sbjct: 625 QSQDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKSNSKKAP 452
Score = 29.6 bits (65), Expect = 2.3
Identities = 13/63 (20%), Positives = 29/63 (45%)
Frame = -1
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGET 100
+S+ EG E E+E++ ++ + +++ DE+D+ ++D+D E
Sbjct: 475 KSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDEE 296
Query: 101 ESE 103
E
Sbjct: 295 NEE 287
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 45.1 bits (105), Expect = 5e-05
Identities = 26/85 (30%), Positives = 40/85 (46%), Gaps = 1/85 (1%)
Frame = +3
Query: 20 SPSSDSTTTTNVNTTTILTTAESSDDNDDE-GPFFDLEFTAPEEEEDGFEETNHKNQQQE 78
S + D NVN + DDNDDE F + E + D E N +
Sbjct: 276 SDTEDDDDDDNVN--------DGEDDNDDEEDEDFSGDDGGEEADSDDDPEANGGGGSDD 431
Query: 79 EEKDEDDDEDSDDDDDDDSGETESE 103
++ D+D+D+D+D+DD D+ E E +
Sbjct: 432 DDGDDDEDDDNDEDDGDEDDEDEGD 506
Score = 38.9 bits (89), Expect = 0.004
Identities = 24/92 (26%), Positives = 46/92 (49%)
Frame = +3
Query: 13 GGAVVGLSPSSDSTTTTNVNTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNH 72
G V L + T ++ ILT + +D N+ P D F P ++ + T+
Sbjct: 87 GSFVEALVLDTFLATAKSLACFVILTGSLLTDVNNLVSPMVDGRF--PLDQLCKGKHTSE 260
Query: 73 KNQQQEEEKDEDDDEDSDDDDDDDSGETESEF 104
+N+ + +D+DDD++ +D +DD+ E + +F
Sbjct: 261 ENKDASDTEDDDDDDNVNDGEDDNDDEEDEDF 356
Score = 36.6 bits (83), Expect = 0.019
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 10/74 (13%)
Frame = +3
Query: 40 AESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDE-------DDDEDSDDD 92
A ++D+DD+ D E +EE++ F + + ++ E DDD+ DD+
Sbjct: 273 ASDTEDDDDDDNVNDGEDDNDDEEDEDFSGDDGGEEADSDDDPEANGGGGSDDDDGDDDE 452
Query: 93 DDD---DSGETESE 103
DDD D G+ + E
Sbjct: 453 DDDNDEDDGDEDDE 494
Score = 34.7 bits (78), Expect = 0.072
Identities = 19/68 (27%), Positives = 32/68 (46%), Gaps = 2/68 (2%)
Frame = +3
Query: 38 TTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDD--DDD 95
T+ E+ D +D E D E++ D E+ + EE D DDD +++ DD
Sbjct: 252 TSEENKDASDTEDDDDDDNVNDGEDDNDDEEDEDFSGDDGGEEADSDDDPEANGGGGSDD 431
Query: 96 DSGETESE 103
D G+ + +
Sbjct: 432 DDGDDDED 455
>TC212752 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific protein T2
precursor, partial (35%)
Length = 533
Score = 44.7 bits (104), Expect = 7e-05
Identities = 21/55 (38%), Positives = 35/55 (63%)
Frame = +3
Query: 50 GPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEF 104
G D + EEE EE + +++++EEE++E+++E DD + +D GE ESEF
Sbjct: 6 GTSSDKQKQEEEEEPHRMEEDDDESEEEEEEEEEEEEEVWDDWEGEDEGERESEF 170
>TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%)
Length = 609
Score = 44.3 bits (103), Expect = 9e-05
Identities = 21/58 (36%), Positives = 34/58 (58%)
Frame = +3
Query: 59 APEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERS 116
A + +E G E + ++ EE+ DED+++D DDDDDDD E K + +P + R+
Sbjct: 108 AIQGDEFGDLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEEESKTKKKSSAPKKSGRA 281
Score = 42.0 bits (97), Expect = 4e-04
Identities = 20/65 (30%), Positives = 33/65 (50%)
Frame = +3
Query: 62 EEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSDANLS 121
E G E + ++ + +E+ +EDDDED +DDDDDD + + E T S+ +
Sbjct: 105 EAIQGDEFGDLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEEESKTKKKSSAPKKSGRAQ 284
Query: 122 LSPSE 126
L +
Sbjct: 285 LGDGQ 299
Score = 41.6 bits (96), Expect = 6e-04
Identities = 17/45 (37%), Positives = 26/45 (57%)
Frame = +3
Query: 57 FTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETE 101
FT + D F + ++ E+D+D+DE+ DDDDDDD + E
Sbjct: 96 FTGEAIQGDEFGDLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEE 230
Score = 40.8 bits (94), Expect = 0.001
Identities = 20/71 (28%), Positives = 35/71 (49%)
Frame = +3
Query: 56 EFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNER 115
EF E++ED + ++ ++E +EDDD+D DDDDD++ +T+ + +
Sbjct: 123 EFGDLEDDED-----DEDIEEDDDEDEEDDDDDDDDDDDEEESKTKKKSSAPKKSGRAQL 287
Query: 116 SDANLSLSPSE 126
D P E
Sbjct: 288 GDGQQGERPPE 320
>TC227389 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (63%)
Length = 1144
Score = 43.9 bits (102), Expect = 1e-04
Identities = 27/86 (31%), Positives = 44/86 (50%), Gaps = 11/86 (12%)
Frame = +3
Query: 43 SDDNDDEGPFFDLEFTAPEEEEDGFEETNHKN----------QQQEEEKDEDDDEDSDDD 92
SD++D++ P + E PE + + + K Q + E +D++EDSDDD
Sbjct: 387 SDEDDEDVPLINTENGKPETKAEDLKVPESKKAVAKASGSAKQVKVVEPKKDNEEDSDDD 566
Query: 93 -DDDDSGETESEFKITLSPSNNERSD 117
DDDD G ++ E + S S++E D
Sbjct: 567 SDDDDFGSSDEEMEDADSDSDDESGD 644
Score = 33.5 bits (75), Expect = 0.16
Identities = 20/65 (30%), Positives = 34/65 (51%), Gaps = 2/65 (3%)
Frame = +3
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDSDDD--DDDDSGETESEFKITLSPSNNERSDA 118
+ EED ++++ + +E+ ED D DSDD+ D+DD ET + K+ L S +
Sbjct: 540 DNEEDSDDDSDDDDFGSSDEEMEDADSDSDDESGDEDDDEETPPK-KVDLGKKRPNESAS 716
Query: 119 NLSLS 123
+S
Sbjct: 717 KTPVS 731
>TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembly protein,
partial (20%)
Length = 777
Score = 43.5 bits (101), Expect = 2e-04
Identities = 21/50 (42%), Positives = 29/50 (58%), Gaps = 1/50 (2%)
Frame = +2
Query: 57 FTAPEEEEDGFEETNHKNQQQE-EEKDEDDDEDSDDDDDDDSGETESEFK 105
FT + D F + ++ EE D++D+ED DDDDDDD E ES+ K
Sbjct: 47 FTGEAIQGDEFGDLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEEESKTK 196
Score = 41.2 bits (95), Expect = 8e-04
Identities = 20/49 (40%), Positives = 33/49 (66%), Gaps = 1/49 (2%)
Frame = +2
Query: 59 APEEEEDGFEETNHKNQQQEEEKDEDD-DEDSDDDDDDDSGETESEFKI 106
A + +E G E + ++ EE+ DED+ D+D DDDDDDD E++++ K+
Sbjct: 59 AIQGDEFGDLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEEESKTKKKV 205
Score = 40.0 bits (92), Expect = 0.002
Identities = 17/48 (35%), Positives = 31/48 (64%)
Frame = +2
Query: 56 EFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
EF E++ED + ++ ++E +EDDD+D DDDDD++ +T+ +
Sbjct: 74 EFGDLEDDED-----DEDIEEDDDEDEEDDDDDDDDDDDEEESKTKKK 202
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 43.5 bits (101), Expect = 2e-04
Identities = 26/65 (40%), Positives = 36/65 (55%), Gaps = 5/65 (7%)
Frame = +1
Query: 44 DDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKD-----EDDDEDSDDDDDDDSG 98
DD+D+E D E AP+ +D +E +EEE D E DD+D+DDDD+D+
Sbjct: 214 DDDDNEDEEDDDEDDAPDGGDDDDDE------DEEEESDVQRGGEPDDDDNDDDDEDEDE 375
Query: 99 ETESE 103
E E E
Sbjct: 376 EDEEE 390
Score = 41.2 bits (95), Expect = 8e-04
Identities = 23/67 (34%), Positives = 33/67 (48%)
Frame = +1
Query: 37 LTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
L TAE + + D F+ E EEEED +E K++ + K D D+ DDD +D
Sbjct: 436 LVTAEEEEASSD----FEPEENGEEEEEDVDDEDGEKSEAPPKRKRSDKDDSDDDDGGED 603
Query: 97 SGETESE 103
E S+
Sbjct: 604 DDERPSK 624
Score = 40.0 bits (92), Expect = 0.002
Identities = 17/44 (38%), Positives = 26/44 (58%)
Frame = +1
Query: 74 NQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSD 117
+ + EE+ DEDD D DDDDD+ E ES+ + P +++ D
Sbjct: 223 DNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDD 354
Score = 38.1 bits (87), Expect = 0.006
Identities = 28/92 (30%), Positives = 38/92 (40%), Gaps = 29/92 (31%)
Frame = +1
Query: 41 ESSDDNDDE----------GPFFDLE--------FTAPEEEEDGFEETNHKNQQQEEEKD 82
+ +D+DDE G DL TA EEE E +++EE+ D
Sbjct: 337 DDDNDDDDEDEDEEDEEEQGEEADLGTEYLIRPLVTAEEEEASSDFEPEENGEEEEEDVD 516
Query: 83 EDDDE-----------DSDDDDDDDSGETESE 103
++D E D DD DDDD GE + E
Sbjct: 517 DEDGEKSEAPPKRKRSDKDDSDDDDGGEDDDE 612
Score = 36.6 bits (83), Expect = 0.019
Identities = 22/87 (25%), Positives = 39/87 (44%)
Frame = +1
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGET 100
E DD DD D + EEEE + + ++ DED+DE+ D+++ + +
Sbjct: 235 EEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDEDEE-DEEEQGEEADL 411
Query: 101 ESEFKITLSPSNNERSDANLSLSPSED 127
+E+ I E +A+ P E+
Sbjct: 412 GTEYLIR-PLVTAEEEEASSDFEPEEN 489
Score = 33.9 bits (76), Expect = 0.12
Identities = 17/55 (30%), Positives = 26/55 (46%), Gaps = 12/55 (21%)
Frame = +1
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDS------------DDDDDDDSGETESE 103
++++D +E + + D+DDDED DDDD+DD E E E
Sbjct: 211 DDDDDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDEDE 375
Score = 32.3 bits (72), Expect = 0.36
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 6/51 (11%)
Frame = +2
Query: 52 FFDLEFTAPEEEEDGFEET------NHKNQQQEEEKDEDDDEDSDDDDDDD 96
F DL+ A +E EE + + ++ E +DE D+ DD+DDDD
Sbjct: 50 FLDLKMKATKELSQHVEENEVTVLVDEEEDEEYESEDEVVDDGEDDEDDDD 202
>TC216024 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F23H14.7
{Arabidopsis thaliana;} , partial (14%)
Length = 1433
Score = 43.1 bits (100), Expect = 2e-04
Identities = 59/261 (22%), Positives = 101/261 (38%), Gaps = 9/261 (3%)
Frame = +2
Query: 59 APEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSDA 118
A E +E + T HK K D DSD+D DD+S +T R +
Sbjct: 293 AAETKERKLKATKHK-------KRSGSDSDSDNDSDDESRKTSKR------SHRKHRKHS 433
Query: 119 NLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEPNNKPQFTASLLKSATKFRVFMS 178
+ E K + K SS ++ ++ SSSE + + K + K R S
Sbjct: 434 HYDSGDHEKRKEKSSKRKTKKSSSETSDYDDFESSSEEERRKK------KQSKKLRDRGS 595
Query: 179 GLKKPKTDSVLE-----KKQHKPER-IKFKVEEVPIV---SLFTRDNSSKNSNKSQTNQN 229
+DS+ + K+ HK +R KF + SL + N K+S + + +++
Sbjct: 596 RSDSSDSDSIADELSKRKRHHKRQRPSKFSGSDYSTEEGDSLVRKRNHGKHSKRHRRSES 775
Query: 230 QKVEHESSLSPLEEKRFSKEIVMHKYLKMVKPLYIRVSRRYSHGDKSSPSEKVEAETATE 289
+ + S +P +K K HK V+ +SHG +S
Sbjct: 776 SESDLSSDDAP-RKKGHRKHQKHHKRSHNVELRSSDSDYHHSHGQRS------------R 916
Query: 290 GETVAENEENNAKTQSQKQGN 310
+++ EN E+ +K K+ +
Sbjct: 917 SKSLEENSEDESKRSMHKKSD 979
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.304 0.123 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,478,358
Number of Sequences: 63676
Number of extensions: 176660
Number of successful extensions: 5356
Number of sequences better than 10.0: 374
Number of HSP's better than 10.0 without gapping: 2111
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3426
length of query: 374
length of database: 12,639,632
effective HSP length: 99
effective length of query: 275
effective length of database: 6,335,708
effective search space: 1742319700
effective search space used: 1742319700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC140913.4


